Training
Training Modules
Useful Links
Observation Time
Joint Trainings
Additional Trainings
CryoEM Learning
CryoEM Course 2025
Training on all instruments is mandatory to ensure functional safe operation of our highly specialized equipment. Separate training is necessary for every instrument including each microscope. Our training modules aim to provide users with the necessary skills to acquire electron microscopy data. Users will be graded according to their level of experience for each respective microscope. Records are kept with the Facility Staff. Pre-requisite for all training is the “New User Orientation”
Training Modules
Sample Preparation training
1. Negative stain and sample preparation training
Description: Training for general negative stain electron microscopy sample preparation, including theory, handling of grids, glow discharging (GlowQube, Harrick Plasma Cleaner)
Prerequisites: New User Orientation
Duration: typically one training session (1-2h) with EM facility staff, depending on requirements and aptitude
Occupancy: Maximum 2 people
Supplies provided by facility: Negative stain reagents depending on requirement, filter paper
Supplies to be provided by users: Prepared sample, grid box, continuous carbon grids, tweezers
2. Carbon Coater
Description: Training to use the Leica ACE600 carbon coater, practical application, recipe creation
Prerequisites: New User Orientation
Duration: typically one training session (1h) with EM facility staff
Occupancy: Maximum 2 people
Supplies provided by facility: Carbon rods, Mica
Supplies to be provided by users: Formvar coated or other grids
3. Vitrobot plunge freezer training
Description: Training to use the Vitrobot plunge freezer, including cryo-sample preparation theory, setup, practical application, use of GloQube (or Harrick plasma cleaner )
Prerequisites: New User Orientation
Duration: up to two training sessions (2h each) with EM facility staff (one training session, if prior training on cryo-plungers was completed)
Occupancy: Maximum 1 person
Supplies provided by facility: plunging tweezers
Supplies to be provided by users: Whatman Paper Grade 1 Ø 55mm, cryo-optimised sample, cryo-grid box with lid, cryo-grids according to user requirements
4. Manual Plunge freezer
Description: Training to use the manual plunge freezer, including cryo-sample preparation theory, setup, practical application, use of GloQube (or Harrick plasma cleaner)
Prerequisites: New User Orientation
Duration: up to 2 separate, 2-hour sessions with EM facility staff, 1 hour if prior training on cryo-plungers was completed
Occupancy: Maximum 1 person
Supplies provided by facility: Filter paper (Whatman #1), plunging tweezers (for training only)
Supplies to be provided by users: cryo-optimised sample, cryo-grid box with lid, cryo-grids according to user requirements, plunging tweezers (after training)
5. Leica GP2 Plunge freezer
Description: Training to use the Leica GP2 Plunge freezer, including cryo-sample preparation theory, setup, practical application, use of GloQube (or Harrick plasma cleaner)
Prerequisites: New User Orientation
Duration: up to two training sessions (2h each) with EM facility staff(one training session, if prior training on cryo-plungers was completed)
Occupancy: Maximum 1 person
Supplies provided by facility: plunging tweezers
Supplies to be provided by users: Whatman Paper Grade 1 Ø 55mm, cryo-optimised sample, cryo-grid box with lid, cryo-grids according to user requirements
6. Chameleon Plunge Freezer Training TBC
Description: Training to use the SPT Labtech Chameleon for preparing grids.
Prerequisites: New User Orientation. It is helpful if you have already imaged the sample on Vitrobot-prepared grids.
Duration: two training sessions (2h/each). Training with just 1 session may be possible after observing another training session.
Occupancy: 1 person. Up to 2 people may observe.
Supplies provided by the Marlovits group: Instrument consumables except grids.
Supplies needed: FEI autogrid box, nanowire grids, cryo-optimsed sample.
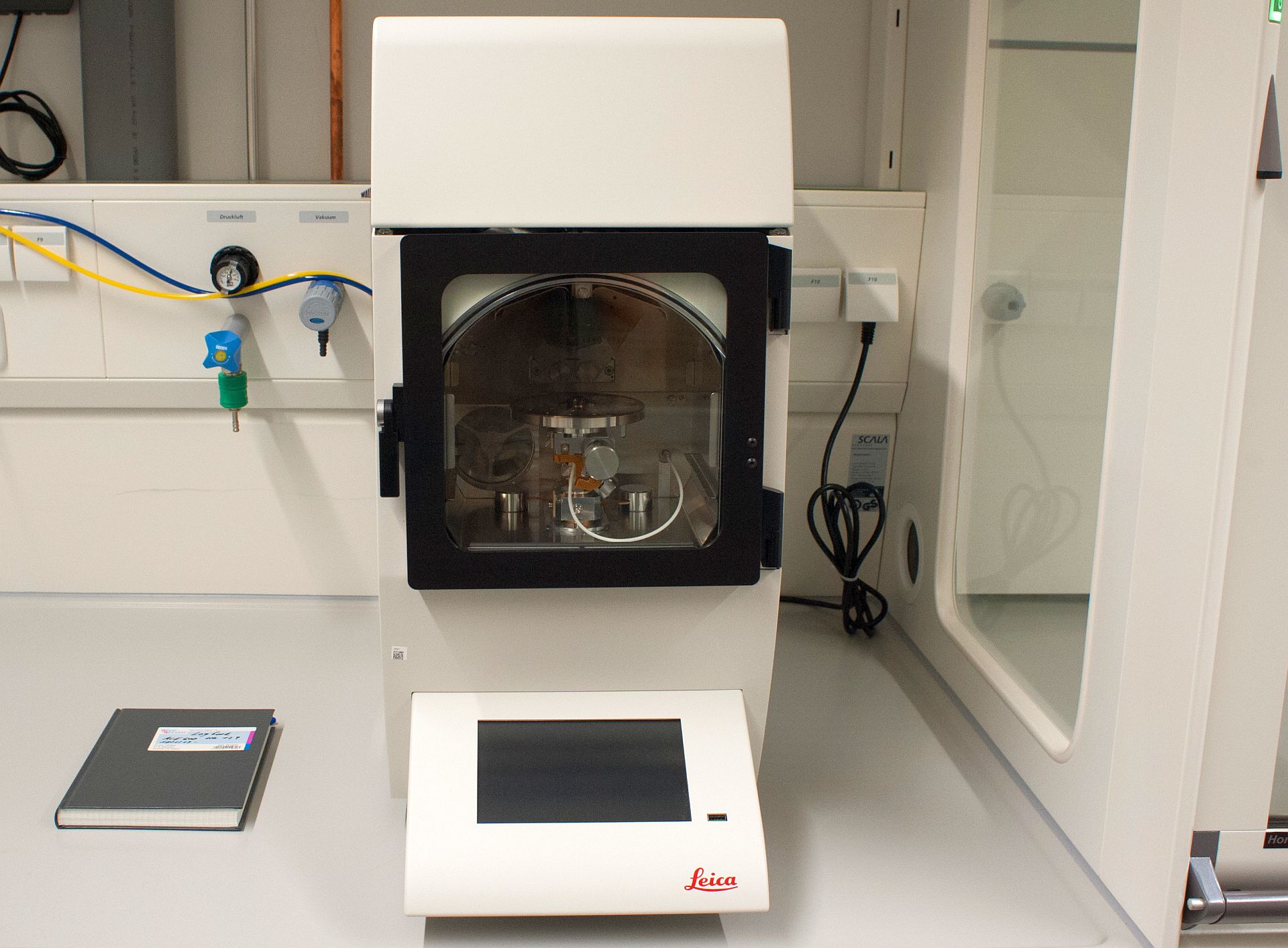
7. High Pressure Freezer Leica EM ICE
Description: Training on how to use the Leica EM ICE High Pressure Freezer (HPF) to prepare tissue or organoid samples and samples using the waffle method.
Prerequisites: Please contact ALFM staff for training
Microscope training
8. Autogrid Clipping
Description: Training to prepare cryoEM autogrids
Prerequisites: New User Orientation, prepared cryo grids
Duration: typically one training session (2h), 1 hour demonstration by EM facility staff and 1 hour practice
Occupancy: Maximum 1 person
Supplies provided by facility: Clipping tools, short term storage for cryo-grids (max 2 weeks)
Supplies to be provided by users: autogrid cryo-box, vitrified cryoEM grids, autogrid rings and C-clips
Microscope training
9. Talos L120
Description: Training and hands-on practical safe use of the scope for screening negative stain grids. Theory, microscope hardware, loading samples, aligning the scope, imaging strategies, focusing, aberrations, troubleshooting. Training sample provided or own sample.
Prerequisites: New User Orientation, negative stain and sample preparation training
Duration: typically two training sessions (2h each) with EM facility staff, more if necessary
Observation session with independent users: 4 hours
Final approval and sign off after 10-20 hours of supervised instrument time logged and demonstration of proficiency
Occupancy: 1-2 people
Supplies needed: neg stained grids (user sample or training sample)
10. Talos Arctica
Description: Training and hands-on practical use of Arctica for screening cryo-grids with Falcon IVi. Training covers theory, microscope hardware, aligning the scope, imaging strategies, focusing, aberrations, troubleshooting.
Prerequisites: cryo grid sample preparation training, autogrid clipping proficiency
Duration:typically four training sessions (4h each) with EM facility staff, more if necessary. Observation sessions with independent users: min 2 x 4 hours.
Final approval and sign off after ~100 hours of supervised instrument time logged by facility staff and demonstration of proficiency
Occupancy: 1-2 people
Supplies needed: vitrified autogrids
11. Titan Krios– EPU/TOMO
Description: Training and hands-on practical use of Krios for setting up high-resolution energy filtered data collection (npEFTEM) with K3 (EPU and TOMO). Theory, microscope hardware, aligning the scope, imaging strategies, focusing, aberrations, troubleshooting.
Prerequisites: cryo grid sample preparation training and autogrid clipping proficiency
Duration:typically two training sessions (6-8h each) with EM facility staff, more if necessary.
Observation sessions with independent users: min 2 x 4 hours
Final approval and sign off after ~100 hours of supervised instrument time logged by facility staff and demonstration of proficiency
Occupancy: 1-2 people
Supplies needed: screened and vitrified autogrids with proven quality
12. Titan Krios - Tomography
Description: Theory and hands-on training of tomography data acquisition in batch with TFS Tomography software and/or serialEM at the Krios microscopes. Includes specific alignment steps and dose calibration of the K3 direct detector for tilt series acquisition. Before introduction to serialEM, principles of tomography software should be understood with the more basic TFS Tomo.
Prerequisites: cryo grid sample preparation training and autogrid clipping proficiency
Duration: typically two training sessions (6-8h each) with EM facility staff, more if necessary
Observation session with independent users: 2 x 4 hours, probably more will be needed
Final approval and sign off after ~100 hours of supervised instrument time logged by facility staff and demonstration of proficiency
Occupancy: 1-2 people
Supplies needed: vitrified autogrids
13. Aquilos FIB SEM
Description: Theory and hands-on training of preparing FIB-thinned lamellae from cellular samples including preparation steps for cryo work on the scope, using microscope software XTUI and additional software MAPS and AutoLamella for autonomous lamella preparation
Prerequisites: sample preparation training of cellular/other suitable samples, plunge freezing and autogrid clipping proficiency.
Duration: typically two training sessions (6-8h each) with EM facility staff, more if necessary
Observation session with independent users: 2 x 4 hours
Final approval and sign off after ~100 hours of supervised instrument time logged by facility staff and demonstration of proficiency
Occupancy: 1-2 people
Supplies needed: vitrified FIB-milling autogrids
14. Arctis Cryo-Plasma FIB SEM
Description: Theory and hands-on training of preparing FIB-milled lamellae from cellular samples or specifically prepared waffle grids, identification of suitable milling areas through fluorescence, use of web-based software application WebUI for remote access, or Maps/AutoTEM workflow
Prerequisites: sample preparation training of cellular samples/waffle grids, plunge freezing and autogrid clipping proficiency
Duration: typically two 2-day training sessions (6-8h/day) with EM facility staff, more if necessary
Observation session with independent users: 2 x 4 hours
Final approval and sign off after ~100 hours of supervised instrument time logged by facility staff and demonstration of proficiency
Occupancy: 1-2 people
Supplies needed: vitrified cellular sample on FIB-milling autogrids or high pressure frozen waffle grids
Links for our Users
Training manuals can be found here. Here is a list of necessary supplies.
Observation Time
In order to acquire proficiency on any instrument, users are encouraged to join experienced user sessions. This allows new users to see different imaging strategies specific to their requirements.
Joint Trainings
Joint trainings complement individual training modules and will allow new users with similar experiences and requirements to spend time together on the microscope, exchange knowledge and foster learning.
Additional Training
After you have completed your initial training, observation time and group training - you may require additional training. We also offer refresher training to ensure that you know how to correctly operate the equipment, and inform you of any recent updates in operating procedures. For additional trainings, please contact cryoem@cssb-hamburg.de, to discuss further needs.
CryoEM Learning
To get started in CryoEM, we recommend to look into the following excellent online resources:
-
We recommend to subscribe to these mailing lists:
- 3DEM email list - community news, technical discussion, data processing, job advertisement
- CCPEM email list - single particle data acquisition and processing
- SerialEM Discussion group - troubleshooting feedback by the developers
-
For a short and general introduction to CryoEM, see Eva Nogales’ iBiology seminar
-
For a more detailed introduction, see the lecture series on EM given at the MRC Laboratory of Molecular Biology - LMB
-
See Caltech’s page for a complete 14-hour theoretical course on EM basics, tomography and SPA in easy to follow video format: Grant Jensen's online class „Getting started in cryo-EM"
-
Jensen’s course together with a practical video tutorial on SPA by Thermo Fisher Scientific and the NIH, the em-learning platform has collected all the knowledge in one place. It is free, but registration is required
-
Lecture series by the SBGrid consortium on relevant software packages for data processing
-
The National Resource for Automated Molecular Microscopy (NRAMM) offers a helpful list of Lectures from workshops
-
Workshops on acquisition and processing software can be found here:
CryoEM Course 2025
The Multi-User cryoEM facility is holding the sixth cryoEM full-day workshop this year between February 10th and 21st, 2025. The workshop targets M.Sc students in Life Sciences as well as beginners in the cryo-EM field and provides an overview of the different modalities of electron cryo-microscopy from single particle analysis (cryo-SPA) to tomography (cryo-ET). Participants will gain knowledge of microscope hardware, sample preparation techniques, image formation, and various modes of scope operation. An integral part of the course are (live) demonstrations on the microscopes and a hands-on introduction to image processing for SPA utilizing cryoSPARC. The course will finish with research talks from PhDs and Postdocs, and M.Sc. students will present a cryoEM or cryoET paper. After successful completion of this course, participants will be able to explain the modalities cryo-SPA and cryo-tomography and evaluate a standard SPA data set from micrographs to a 3D model. More details to follow.
Organizers: Carolin Seuring, Ulrike Laugks, Cornelia Cazey, Katharina Jungnickel and Kay Grünewald
Programme will be available soon! https://www.uni-hamburg.de/en/technologieplattformen/technologien/kryo-elektronenmikroskopie/kurse-workshops.html
Publications
Any use of the resources at the Multi-User CryoEM facility at CSSB requires acknowledgement in publications as follows: “Part of this work was performed at the Multi-User CryoEM Facility at the Centre for Structural Systems Biology, Hamburg, supported by the Universität Hamburg and DFG grant numbers (INST 152/772-1|152/774-1|152/775-1|152/776-1|152/777-1 FUGG), the Federal Ministry of Education and Research (BMBF) and the DLR Projektträger (project SEEK 01KX2220). Any assistance provided by staff should be acknowledged. Facility staff members who have acquired and/or interpreted data on your behalf should be invited to be co-authors on the publication, as is usual practice.
2024
Ekeberg T, Assalauova D, Bielecki J, Boll R, Daurer BJ, Eichacker LA, Franken LE, Galli DE, Gelisio L, Gumprecht L, Gunn LH, Hajdu J, Hartmann R, Hasse D, Ignatenko A, Koliyadu J, Kulyk O, Kurta R, Kuster M, Lugmayr W, Lübke J, Mancuso AP, Mazza T, Nettelblad C, Ovcharenko Y, Rivas DE, Rose M, Samanta AK, Schmidt P, Sobolev E, Timneanu N, Usenko S, Westphal D, Wollweber T, Worbs L, Xavier PL, Yousef H, Ayyer K, Chapman HN, Sellberg JA, Seuring C, Vartanyants IA, Küpper J, Meyer M, Maia FRNC (2024) Observation of a single protein by ultrafast X-ray diffraction. Light Sci Appl. 13(1):15. doi: 10.1038/s41377-023-01352-7.
Gersteuer, F, Morici, M, Gabrielli, S, Fujiwara, K, Safdari, HA, Paternoga, H, Bock, LV, Chiba, S, Wilson, DN, (2024). The SecM arrest peptide traps a pre-peptide bond formation state of the ribosome. Nat. Commun. 15, 2431. https://doi.org/10.1038/s41467-024-46762-2
Goradia N, Werner S, Mullapudi E, Greimeier S, Bergmann L, Lang A, Mertens H, Weglarz A, Sander S, Chojnowski G, Wikman H, Ohlenschläger O, von Amsberg G, Pantel K, Wilmanns M, (2024). Master corepressor inactivation through multivalent SLiM-induced polymerization mediated by the oncogene suppressor RAI2. Nat. Commun. 15. https://doi.org/10.1038/s41467-024-49488-3
Gustavsson, E, Grünewald, K, Elias, P, Hällberg, BM, (2024). Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM. Nucleic Acids Res. 52, 7292–7304. https://doi.org/10.1093/nar/gkae374
Hofstadter, WA, Cook, KC, Tsopurashvili, E, Gebauer, R, Prazak, V, Machala, EA, Park, JW, Gruenewald, K, Quemin, ERJ, Cristea, IM, (2024). Infection-induced peripheral mitochondria fission drives ER encapsulations and inter-mitochondria contacts that rescue bioenergetics. Nat. Commun. 15. https://doi.org/10.1038/s41467-024-51680-4
Jungnickel, KEJ, Guelle, O, Iguchi, M, Dong, W, Kotov, V, Gabriel, F, Debacker, C, Dairou, J, McCort-Tranchepain, I, Laqtom, NN, Chan, SH, Ejima, A, Sato, K, Massa Lopez, D, Saftig, P, Mehdipour, AR, Abu-Remaileh, M, Gasnier, B, Low, C, Damme, M, (2024). MFSD1 with its accessory subunit GLMP functions as a general dipeptide uniporter in lysosomes. Nat. Cell Biol. 26, 1047–1061. https://doi.org/10.1038/s41556-024-01436-5
Liu, J, Corroyer-Dulmont, S, Prazak, V, Khusainov, I, Bahrami, K, Welsch, S, Vasishtan, D, Obarska-Kosinska, A, Thorkelsson, SR, Grunewald, K, Quemin, ERJ, Turonova, B, Locker, JK, (2024). The palisade layer of the poxvirus core is composed of flexible A10 trimers. Nat. Struct. Mol. Biol. 31, 1105–1113. https://doi.org/10.1038/s41594-024-01218-5
Morici, M, Gabrielli, S, Fujiwara, K, Paternoga, H, Beckert, B, Bock, LV, Chiba, S, Wilson, DN, (2024). RAPP-containing arrest peptides induce translational stalling by short circuiting the ribosomal peptidyltransferase activity. Nat. Commun. 15, 2432. https://doi.org/10.1038/s41467-024-46761-3
Ott, F, Jonsson, MR, Grunewald, K, Hellert, J, (2024). Preparation of Bunyavirus-Infected Cells for Electron Cryo-Tomography. Methods Mol. Biol. 2824, 221–239. https://doi.org/10.1007/978-1-0716-3926-9_15
Prazak, V, Mironova, Y, Vasishtan, D, Hagen, C, Laugks, U, Jensen, Y, Sanders, S, Heumann, JM, Bosse, JB, Klupp, BG, Mettenleiter, TC, Grange, M, Grunewald, K, (2024). Molecular plasticity of herpesvirus nuclear egress analysed in situ. Nat. Microbiol. 9, 1842–1855. https://doi.org/10.1038/s41564-024-01716-8
Rutten, M, Lang, L, Wagler, H, Lach, M, Mucke, N, Laugks, U, Seuring, C, Keller, TF, Stierle, A, Ginn, HM, Beck, T, (2024). Assembly of Differently Sized Supercharged Protein Nanocages into Superlattices for Construction of Binary Nanoparticle-Protein Materials. ACS Nano 18, 25325–25336. https://doi.org/10.1021/acsnano.4c09551
Williams, HM, Thorkelsson, SR, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, ERJ, Rosenthal, M, (2024). Structural snapshots of phenuivirus cap-snatching and transcription. Nucleic Acids Res. 52, 6049–6065. https://doi.org/10.1093/nar/gkae330
2023
Jungnickel KEJ, Guelle O, Iguchi M, Dong W, Kotov V, Gabriel F, Debacker C, Dairou J, McCort-Tranchepain I, Laqtom NN, Ham Chan D, Ejima A, Sato K, Massa López D, Saftig P, Reza Mehdipour A, Abu-Remaileh M, Gasnier B, Löw C, Damme M (2023) MFSD1 in complex with its accessory subunit GLMP functions as a general dipeptide uniporter in lysosomes bioRxiv 570541. https://doi.org/10.1101/2023.12.15.570541
Custódio TF, Killer M, Yu D, Puente V, Teufel DP, Pautsch A, Schnapp G, Grundl M, Kosinski J, Löw C (2023) Molecular basis of TASL recruitment by the peptide/histidine transporter 1, PHT1. Nat Commun. 14(1):5696. doi: 10.1038/s41467-023-41420-5.
Blanchet CE, Round A, Mertens HDT, Ayyer K, Graewert M, Awel S, Franke D, Dörner K, Bajt S, Bean R, Custódio TF, de Wijn R, Juncheng E, Henkel A, Gruzinov A, Jeffries CM, Kim Y, Kirkwood H, Kloos M, Knoška J, Koliyadu J, Letrun R, Löw C, Makroczyova J, Mall A, Meijers R, Pena Murillo GE, Oberthür D, Round E, Seuring C, Sikorski M, Vagovic P, Valerio J, Wollweber T, Zhuang Y, Schulz J, Haas H, Chapman HN, Mancuso AP, Svergun D (2023) Form factor determination of biological molecules with X-ray free electron laser small-angle scattering (XFEL-SAS). Commun Biol. 6(1):1057. doi: 10.1038/s42003-023-05416-7.
Ferreira JL, Prazak V, Vasishtan D, Siggel M, Hentzschel F, Binder AM, Pietsch E, Kosinski J, Frischknecht F, Gilberger TW, Grunewald K (2023) Variable microtubule architecture in the malaria parasite. Nat Commun 14: 1216 doi: 10.1038/s41467-023-36627-5
Seyfert CE, Porten C, Yuan B, Deckarm S, Panter F, Bader CD, Coetzee J, Deschner F, Tehrani KHME, Higgins PG, Seifert H, Marlovits TC, Herrmann J, Muller R (2023) Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering. Angew Chem Int Edit 6210.1002/anie.202214094
Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grunewald K, Quemin ERJ, Rosenthal M (2023) Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Research 51: 1424-1442 doi: 10.1093/nar/gkac1249
2022
Ekeberg T, Assalauova D, Bielecki J, Boll R, Daurer BJ, Eichacker LA, Franken LE, Galli DE, Gelisio L, Gumprecht L, Gunn LH, Hajdu J, Hartmann R, Hasse D, Ignatenko A, Koliyadu J, Kulyk O, Kurta R, Kuster M, Lugmayr W, Lübke J, Mancuso AP, Mazza T, Nettelblad C, Ovcharenko Y, Rivas DE, Samanta A K, Schmidt P, Sobolev E, Timneanu N, Usenko S, Westphal D, Wollweber T, Worbs L, Xavier PL, Yousef H, Ayyer K, Chapman HN, Sellberg JA, Seuring C, Vartanyants IA, Küpper J, Meyer M, Maia FRNC (2022) Observation of a single protein by ultrafast X-ray diffraction. Bioarxiv, doi: https://doi.org/10.1101/2022.03.09.483477
Franken, L.E., Rosch, R., Laugks, U., and Grünewald, K. (2022). Protocol for live-cell fluorescence-guided cryoFIB-milling and electron cryo-tomography of virus-infected cells, STAR Protocols, Volume 3, Issue 4, 101696, https://doi.org/10.1016/j.xpro.2022.101696.
Niebling S, Veith K, Vollmer B, Lizarrondo J, Burastero O, Schiller J, Struve García A, Lewe P, Seuring C, Witt S, García-Alai M (2022) Biophysical Screening Pipeline for Cryo-EM Grid Preparation of Membrane Proteins. Front. Mol. Biosci. 9:882288. doi: 10.3389/fmolb.2022.882288
Pazicky S, Alder A, Mertens H, Svergun DI, Gilberger T, Low C (2022) N-terminal phosphorylation regulates the activity of Glycogen Synthase Kinase 3 from Plasmodium falciparum. Biochem J 10.1042/BCJ20210829
Killer M, Finocchio G, Mertens HDT, Svergun DI, Pardon E, Steyaert J, Loew C (2022) Cryo-EM Structure of an Atypical Proton-Coupled Peptide Transporter: Di- and Tripeptide Permease C. Front Mol Biosci 9ARTN 91772510.3389/fmolb.2022.917725
Bücker R, Seuring C, Cazey C, Veith K, García-Alai M, Grünewald K, Landau M (2022) The Cryo-EM structures of two amphibian antimicrobial cross-β amyloid fibrils. Nat Commun. 13(1):4356. doi: 10.1038/s41467-022-32039-z
Villalta A, Schmitt A, Estrozi LF, Quemin ER, Alempic JM, Lartigue A, Prazak V, Belmudes L, Vasishtanm D, Colmant AM, Honore FA, Coute Y, Grunewald K, Abergel C (2022) The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield. Elife 11ARTN e77607 doi: 10.7554/eLife.77607
Wald J, Fahrenkamp D, Goessweiner-Mohr N, Lugmayr W, Ciccarelli L, Vesper O, Marlovits TC (2022) Mechanism of AAA plus ATPase-mediated RuvAB-Holliday junction branch migration. Nature 609: 630-+ doi: 10.1038/s41586-022-05121-1
Yuan B, Portaliou AG, Parakra R, Smit JH, Wald J, Li YC, Srinivasu B, Loos MS, Dhupar HS, Fahrenkamp D, Kalodimos CG, van Hoa FD, Cordes T, Karamanou S, Marlovits TC, Economou A (2021) Structural Dynamics of the Functional Nonameric Type III Translocase Export Gate. Journal of Molecular Biology 433ARTN 16718810.1016/j.jmb.2021.167188
2021
Prazak V, Grunewald K, Kaufmann R (2021) Correlative super-resolution fluorescence and electron cryo-microscopy based on cryo-SOFI. Methods Cell Biol 162: 253-271 doi: 10.1016/bs.mcb.2020.10.021
Bunduc CM, Fahrenkamp D, Wald J, Ummels R, Bitter W, Houben ENG, Marlovits TC (2021) Structure and dynamics of a mycobacterial type VII secretion system. Nature 593: 445-448 doi: 10.1038/s41586-021-03517-z
Pfitzner S, Bosse JB, Hofmann-Sieber H, Flomm F, Reimer R, Dobner T, Gruenewald K, Franken LE (2021) Human Adenovirus Type 5 Infection Leads to Nuclear Envelope Destabilization and Membrane Permeability Independently of Adenovirus Death Protein. Int J Mol Sci 22. ARTN 13034. doi: https://doi.org/10.3390/ijms222313034
Kotov V, Lunelli M, Wald J, Kolbe M, Marlovits TC (2021) Helical reconstruction of Salmonella and Shigella needle filaments attached to type 3 basal bodies. Biochem Biophys Rep 27: 101039 doi: 10.1016/j.bbrep.2021.101039
Killer M, Wald J, Pieprzyk J, Marlovits TC, Low C (2021) Structural snapshots of human PepT1 and PepT2 reveal mechanistic insights into substrate and drug transport across epithelial membranes. Sci Adv 7: eabk3259 doi: 10.1126/sciadv.abk3259
Kouba T, Vogel D, Thorkelsson SR, Quemin ERJ, Williams HM, Milewski M, Busch C, Gunther S, Grunewald K, Rosenthal M, Cusack S (2021) Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity. Nat Commun 12: 7018 doi: 10.1038/s41467-021-27305-5
Pfitzner S, Bosse JB, Hofmann-Sieber H, Flomm F, Reimer R, Dobner T, Grunewald K, Franken LE (2021) Human Adenovirus Type 5 Infection Leads to Nuclear Envelope Destabilization and Membrane Permeability Independently of Adenovirus Death Protein. Int J Mol Sci 2210.3390/ijms222313034
Albers S, Beckert B, Matthies M, Mandava C, Schuster R, Seuring C, Riedner M, Sanyal S, Torda A, Wilson D, Ignatova Z (2021) Repurposing tRNAs for nonsense suppression. Nat Commun 12, 3850, https://doi.org/10.1038/s41467-021-24076-x
Kotov V, Mlynek G, Vesper O, Pletzer M, Wald J, Teixeira-Duarte CM, Celia H, Garcia-Alai M, Nussberger S, Buchanan SK, Morais-Cabral JH, Loew C, Djinovic-Carugo K, Marlovits TC (2021) In-depth interrogation of protein thermal unfolding data with MoltenProt. Protein Sci. 30(1):201-217. doi: 10.1002/pro.3986.
Silvester E, Vollmer B, Prazak V, Vasishtan D, Machala EA, Whittle C, Black S, Bath J, Turberfield AJ, Grunewald K, Baker LA (2021) DNA origami signposts for identifying proteins on cell membranes by electron cryotomography. Cell 184: 1110-1121 e1116 doi: 10.1016/j.cell.2021.01.033
Miletic S, Fahrenkamp D, Goessweiner-Mohr N, Wald J, Pantel M, Vesper O, Kotov V, Marlovits TC (2021) Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation. Nat Commun 12: 1546 doi: 10.1038/s41467-021-21143-1
Ayyer K, Xavier PL, Bielecki J, Shen Z, Daurer BJ, Samanta AK, Awel S, Bean R, Barty A, Bergemann M, Ekeberg T, Estillore AD, Fangohr H, Giewekemeyer K, Hunter MS, Karnevskiy M, Kirian RA, Kirkwood H, Kim Y, Koliyadu J, Lange H, Letrun R, Lubke J, Michelat T, Morgan AJ, Roth N, Sato T, Sikorski M, Schulz F, Spence JCH, Vagovic P, Wollweber T, Worbs L, Yefanov O, Zhuang YL, Maia FRNC, Horke DA, Kupper J, Loh ND, Mancuso AP, Chapman HN (2021) 3D diffractive imaging of nanoparticle ensembles using an x-ray laser. Optica 8: 15-23 doi: 10.1364/Optica.410851
2020
Vogel D, Thorkelsson SR, Quemin ERJ, Meier K, Kouba T, Gogrefe N, Busch C, Reindl S, Gunther S, Cusack S, Grunewald K, Rosenthal M (2020) Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Res 48: 5749-5765 doi: 10.1093/nar/gkaa253
Wolff G, Limpens R, Zevenhoven-Dobbe JC, Laugks U, Zheng S, de Jong AWM, Koning RI, Agard DA, Grunewald K, Koster AJ, Snijder EJ, Barcena M (2020) A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369: 1395-1398 doi: 10.1126/science.abd3629
Vollmer B, Prazak V, Vasishtan D, Jefferys EE, Hernandez-Duran A, Vallbracht M, Klupp BG, Mettenleiter TC, Backovic M, Rey FA, Topf M, Grunewald K (2020) The prefusion structure of herpes simplex virus glycoprotein B. Sci Adv 6ARTN eabc172610.1126/sciadv.abc1726
Quemin ERJ, Machala EA, Vollmer B, Prazak V, Vasishtan D, Rosch R, Grange M, Franken LE, Baker LA, Grunewald K (2020) Cellular Electron Cryo-Tomography to Study Virus-Host Interactions. Annu Rev Virol 7: 239-262 doi: 10.1146/annurev-virology-021920-115935
Lunelli M, Kamprad A, Burger J, Mielke T, Spahn CMT, Kolbe M (2020) Cryo-EM structure of the Shigella type III needle complex. PLoS Pathog 16: e1008263 doi: 10.1371/journal.ppat.1008263
Franken LE, Grunewald K, Boekema EJ, Stuart MCA (2020) A Technical Introduction to Transmission Electron Microscopy for Soft-Matter: Imaging, Possibilities, Choices, and Technical Developments. Small: e1906198 doi: 10.1002/smll.201906198
Bunduc CM, Fahrenkamp D, Wald J, Ummels R, Bitter W, Houben ENG, Marlovits TC (2020) Structure and dynamics of the ESX-5 type VII secretion system of Mycobacterium tuberculosis. bioRxiv: 2020.2012.2002.408906 doi: 10.1101/2020.12.02.408906
Beckham KSH, Ritter C, Chojnowski G, Mullapudi E, Rettel M, Savitski MM, Mortensen SA, Kosinski J, Wilmanns M. (2020) Structure of the mycobacterial ESX-5 Type VII Secretion System hexameric pore complex bioRxiv 2020.11.17.387225; doi: https://doi.org/10.1101/2020.11.17.387225.
2019
Nikolaus Goessweiner-Mohr VK, Matthias J. Brunner, Julia Mayr, Jiri Wald, Lucas Kuhlen, Sean Miletic, Oliver Vesper, Wolfgang Lugmayr Samuel Wagner, Frank DiMaio, Susan Lea, Thomas C., Marlovits (2019) Structural control for the coordinated assembly into functional pathogenic type-3 secretion systems. BioArxiv https://doi.org/10.1101/714097
Moser F, Prazak V, Mordhorst V, Andrade DM, Baker LA, Hagen C, Grunewald K, Kaufmann R (2019) Cryo-SOFI enabling low-dose super-resolution correlative light and electron cryo-microscopy. Proc Natl Acad Sci U S A 116: 4804-4809 doi: 10.1073/pnas.1810690116