A Better Understanding of Mitochondrial RNA Processing
Every 30 minutes, a child is born who will develop a mitochondrial disease by age 10. Children with mitochondrial disease suffer from a wide variety of symptoms including strokes, seizures, blindness and deafness and many do not reach adulthood. This disease is often a result of inherited mutations affecting the gene expression from mitochondrial DNA (mtDNA). CSSB group leader, Martin Hällberg’s research focuses on developing an in-depth understanding of the structure and function of mtDNA gene expression within the human cell. A recent paper published by the Hällberg group in Nucleic Acids Research details the structure of a subunit of mitochondrial RNase P, a protein complex essential for mitochondrial RNA processing.
Mitochondria are the power plant of the cell providing energy for all cellular processes within the human body. The mitochondrion stands under dual genetic control. While most of the proteins that work inside mitochondria are encoded in the nuclear genome, 13 proteins critical for power generation are encoded on the mitochondrion’s own genome called the mitochondrial DNA (mtDNA). Impaired gene expression of mtDNA leads to mitochondrial disease. To obtain functional RNA, which can be used in translation, requires extensive processing of the initial RNA transcript. Previous studies have demonstrated that the mitochondrial RNase P (mt-RNase P), a protein complex, initiates this process. In this most recent study, the Hällberg group has focused on understanding MRPP3, one of the three individual proteins (MRPP1-3) that comprise the human mt-RNase P complex.
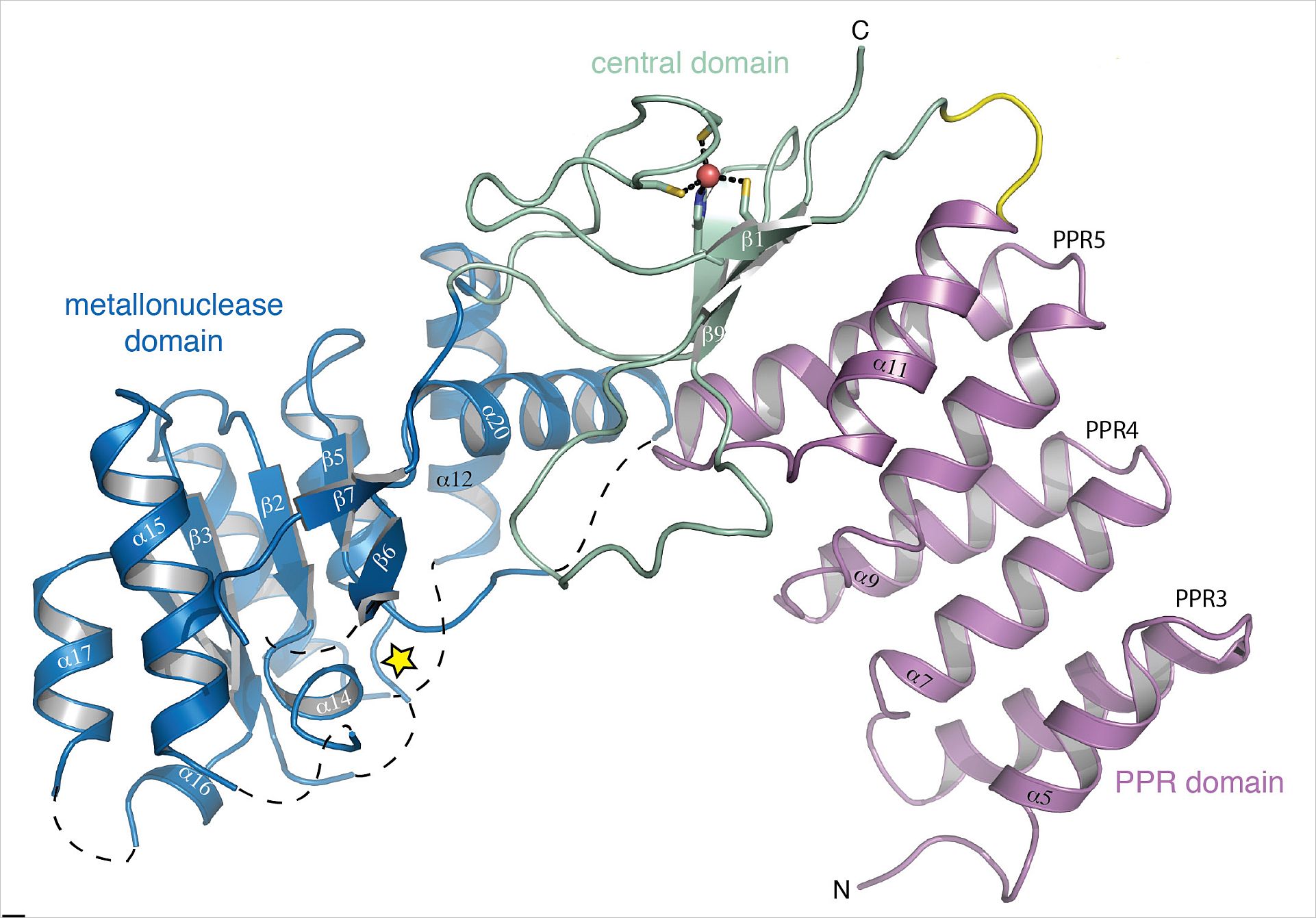
Using the European Molecular Biology Laboratory (EMBL) PETRA III P14 beam line on the DESY campus in Hamburg, Germany, Hällberg and his group were able to determine the crystal structure of MRPP3. The resulting structure shows that the overall structure of MRPP3 alone is strongly distorted and has a dysfunctional active site. “We did not anticipate finding such disarray without the partners MRPP1/2.” explains Hällberg “it was completely unexpected that such large conformational changes would be necessary for activation.” Hällberg and his colleagues were able to prove that MRPP3 does indeed act as the nuclease subunit in the mt-RNase P complex. “Once MRPP3 is joined and stabilized by its complex partners, MRPP1 and MRPP2, and pre-tRNA, order is restored and MRPP3 can act as a nuclease” states Hällberg. These findings bring the Hällberg group a step closer to its goal of understanding the entire structure and function of the mitochondrial RNase P complex.
The work provides a good foundation for the molecular understanding of mitochondrial disease that is caused by defects in initial RNA processing. This in-depth, molecular understanding of mitochondrial gene expression may provide novel avenues for treating human mitochondrial disease. Given the limited existing treatment options currently available for those suffering from this disease, each new piece of the molecular puzzle is crucial.
Original Publication: : http://nar.oxfordjournals.org/content/early/2015/05/07/nar.gkv481.full



