Achieving Atomic Accuracy
Cryo-electron microscopy (cryo-EM) is an emerging method used by structural biologists to further our understanding of molecular structures. Although recent technical developments have led to great improvements in cryo-EM imaging, the development of methods for building accurate models has lagged behind. Last week an international team of scientists including CSSB group leader, Thomas Marlovits*, published a new method for achieving atomic accurate models from cryo-EM data in the peer reviewed journal Nature Methods. “Our protocol generates a clear and accurate atomic structure which places atoms in their precise position even if the original data from the cryo-electron microscopy maps is of a lower resolution” explains Marlovits.
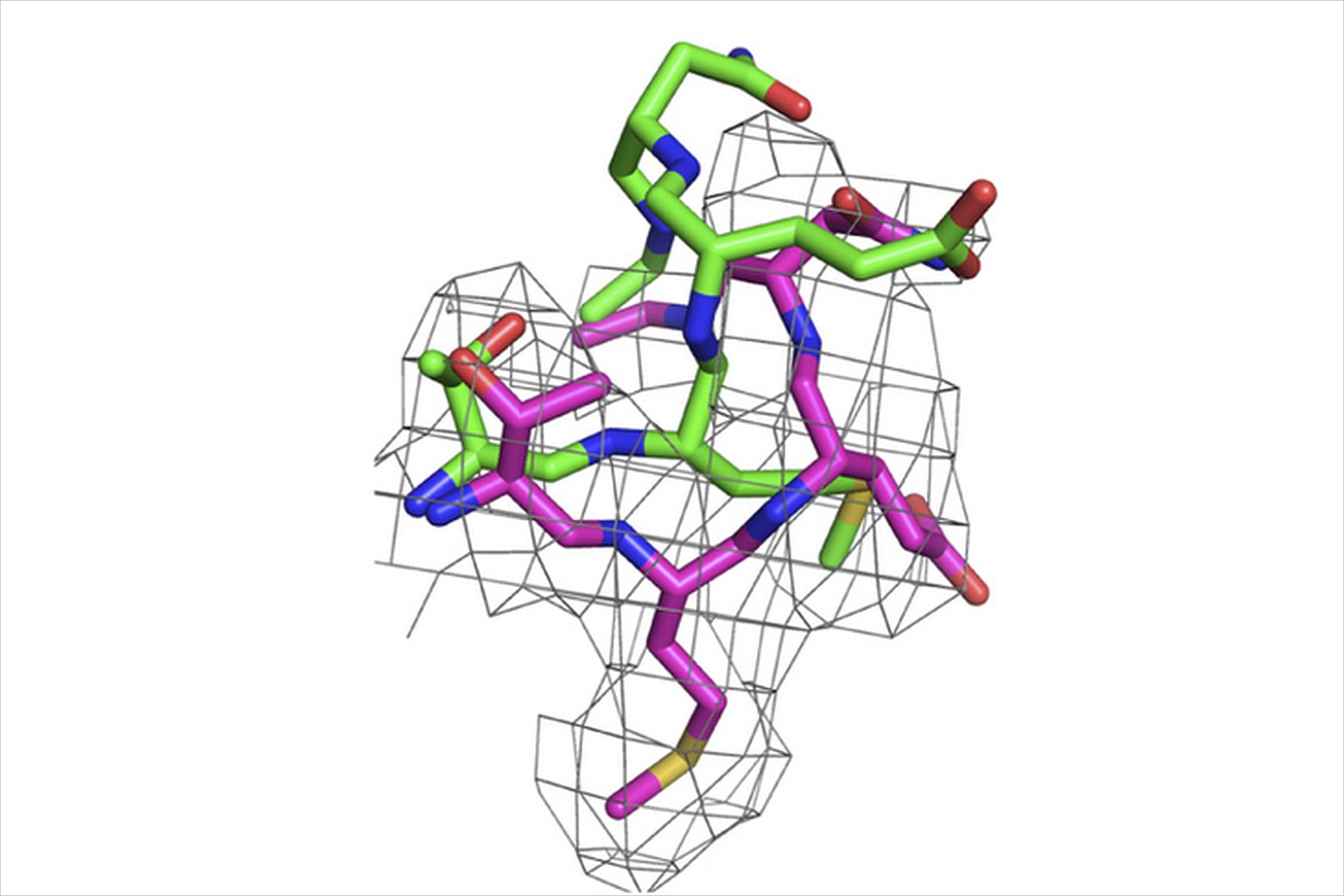
The use of cryo-electron microscopy methods is becoming increasingly popular as it enables the observation of molecules as they are in the cell sometimes even without the need for extensive preparation. However, this means that the molecules will not be sitting perfectly still, resulting in a low resolution image revealing only the overall shape of the molecule. “It’s similar to taking a picture of Formula One race car driving directly in front of you – you get a slightly blurry image” explains Marlovits. “The flexible nature of the sample itself is restricting our ability to capture a high resolution image”.
Scientists have therefore been attempting to generate accurate models by placing atomically resolved structures from other structural biology methods, such as crystallography, into maps showing the density of atoms generated from cryo-EM data. This approach, however, has proven problematic as it often results in a very ambiguous interpretation of the underlying molecular mechanisms. Marlovits and his associates were determined to reduce this ambiguity and their combined efforts resulted in the development of a cryo-EM model refinement protocol which enables the generation of an atomically accurate molecular model even if the data itself is only of near-atomic resolution.
“Given recent technological advances, it is expected that many structures will come out at near-atomic resolution - our protocol can then be used to generate atomic structures” states Marlovits. Used to refine models of protein structures, the protocol could also be applied to other biological molecules. The ability to generate an atomic structure using cryo-EM will not only enable many structural biologists to further their research activities but will also contribute to the growing popularity of the technology.
The CSSB building, to be completed in early 2017, may house up to five cryo-electron microscopes and this technology will play an important role in CSSB’s endeavours to understand the molecular mechanisms of the infection process. “The protocol developed by Thomas Marlovits and his associates has the potential to positively impact the research of many CSSB group leaders” explains Matthias Wilmanns, scientific director of the CSSB, “generating atomically accurate molecular structures from cyro-EM data is an essential step towards advancing our knowledge in the fields of both infection and structural biology.”
* Thomas Malovits is affiliated with the following organizations: Center for Structural Systems Biology (CSSB), University Medical Center Eppendorf-Hamburg (UKE), Hamburg, Germany, Deutsches Elektronen-Synchrotron (DESY), Hamburg, Germany, Institute of Molecular Biotechnology (IMBA), Austrian Academy of Sciences, Vienna, Austria, Research Institute of Molecular Pathology (IMP), Vienna, Austria.
Source Article: Atomic- accuracy models from 4.5-Å cryo-electron microscopy data with density-guided iterative local refinement
Published online 23 February 2015
DOI:10.1038/nmeth.3286



