The Benefit of Longer Peptide Substrates
While it is almost unequivocally accepted that bigger is not always better, longer just might be. A recent paper published by CSSB scientist, Christian Loew, demonstrates that the binding of longer unmodified peptides to the SlyD enzyme results in higher affinity and activity measurement than binding it to a chemically modified tetrapeptide. Using this longer substrate length enabled Loew and his group to present a series of SlyD structures from the organism Thermus thermophilus which reveal important clues about substrate binding and the catalytic mechanism of the enzyme.
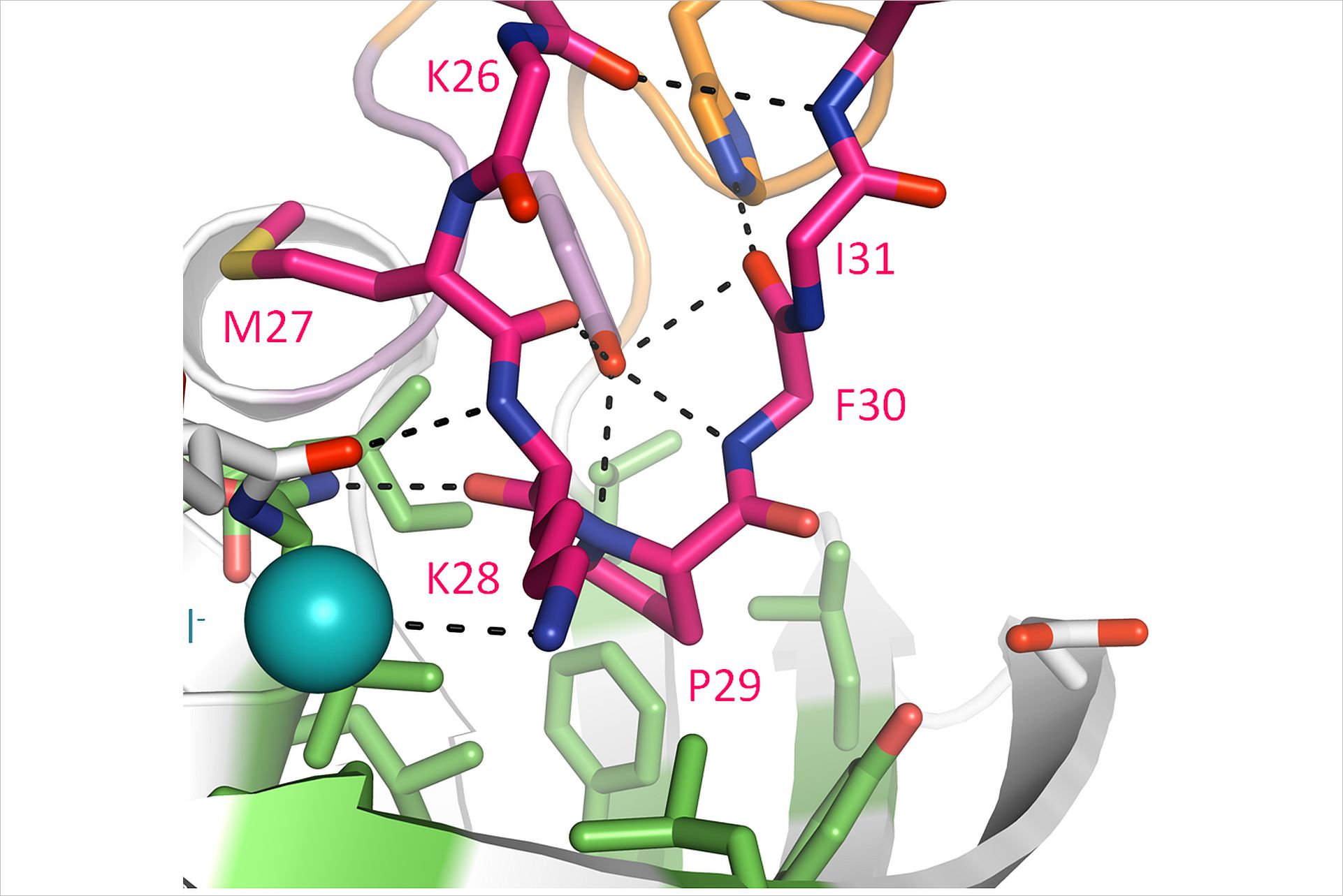
SlyD is a folding helper protein that accelerates the cis/trans isomerization of proline peptide bonds which are important for the correct folding of proteins. Loew has been studying this enzyme since his days as a PhD student at the University of Halle-Wittenberg where he contributed to the structure determination of the substrate free enzyme. He then continued to study this protein as a post-doc attempting to understand on a molecular level how these enzymes bind their substrates and catalyse the isomerization reaction. Initially, he was unable to identify substrates suitable for crystallization studies due to their low affinity and had all but given up on finding more information about SlyD. A contaminant in a membrane protein preparation led to the structure determination of a homologous protein from E. coli and the identification of new substrates.*
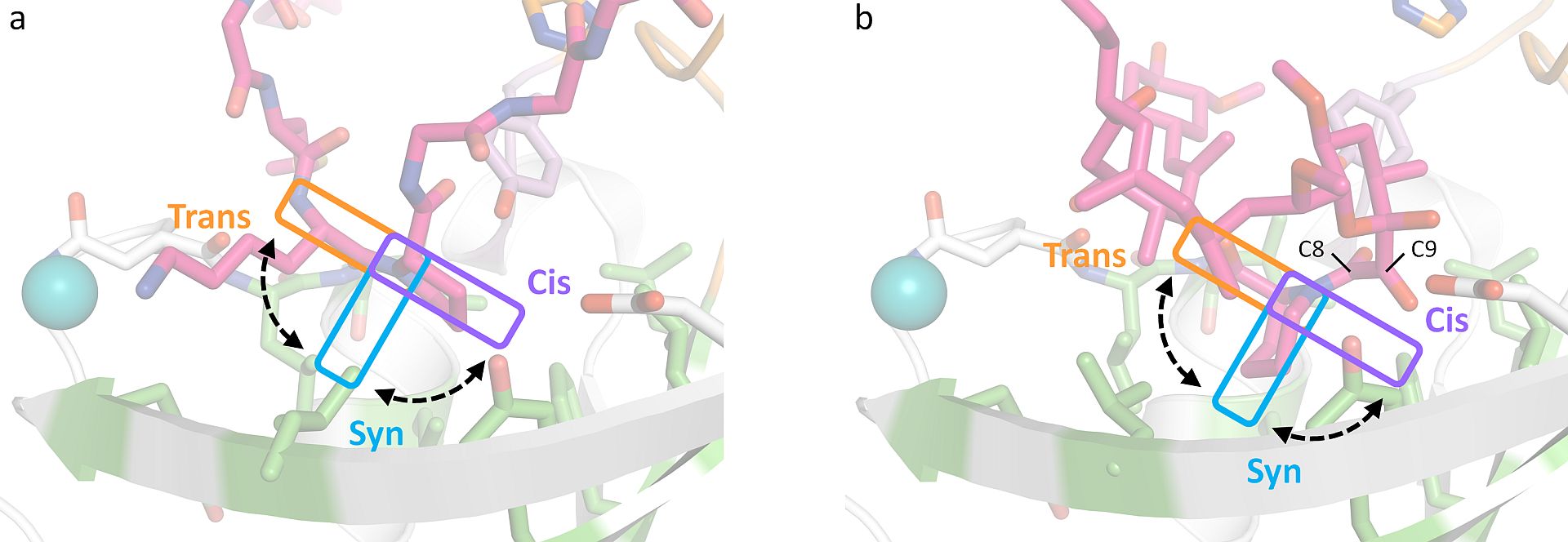
Out of curiosity Loew added these longer peptides to the thermophilic SlyD protein and to his great surprise they bound with high affinity. Crystals of various SlyD-peptide complexes could be grown and their structures determined using X-ray crystallography. He also teamed up with colleagues at the University of Lund and used NMR spectroscopy to analyse the activity towards these new substrates. The results revealed that the substrates bind in a highly adaptable fashion to SlyD and the enzyme displays a 100 - 1000 fold higher activity towards these substrates compared to the conventionally used tetrapeptides. Furthermore, the obtained structures allowed the researchers to propose a novel general model for the catalytic mechanism of prolyl isomerases of the FKBP class that involves C-terminal rotation around the peptidyl-prolyl bond mediated by stabilization of the twisted transition state in the hydrophobic binding site. Loew hopes that his experience will encourage other scientists to consider using longer peptides as substrates in their assays when examining proline isomerase reactions. “These substrate mimics are more authentic,” he explains.
*(ref: PMID: 22735173 FASEB J. 2012 Oct;26(10):4003-13. doi: 10.1096/fj.12-208397. Epub 2012 Jun 26. High-resolution insights into binding of unfolded polypeptides by the PPIase chaperone SlpA. Quistgaard EM1, Nordlund P, Löw C.)
Source Article:
ref: PMID: 27664121 BMC Biol. 2016 Sep 23;14(1):82. doi:10.1186/s12915-016-0300-3. Molecular insights into substrate recognition and catalytic mechanism of the chaperone and FKBP peptidyl-prolyl isomerase SlyD.Quistgaard EM, Weininger U, Ural-Blimke Y, Modig K, Nordlund P, Akke M, Löw C



