Catalysing Collaboration
CSSB Invests 420K Euro in Seed Projects
CSSB will be investing 420K over the course of two years in three seed projects. The funding is intended to serve as catalyst for collaboration among group leaders from different CSSB partner institutions. Financed from the CSSB Director’s fund, the seed projects aim to promote the development of young scientists by supporting the development and adaptation of innovative methods and approaches that have a wider impact on the research performed within CSSB.
“CSSB was established to enable scientists to move beyond the boundaries of individual institutions and research areas,” explains CSSB Scientific Director Chris Meier “these three projects embody this goal and will help propel CSSB to the forefront of integrative and innovative infection biology research.”
Below is short description of each of the three seed projects:
Research Groups: Jan Kosinski (EMBL) and Michael Kolbe (HZI)
Structural analysis of Type 3 Secretion System from Shigella flexneri as a seed for future integrative structure determination projects at CSSB
The T3SS is a membrane-embedded multiprotein complex essential for the translocation of pathogenicity factors into host cells during infection. This project aims to determine the integrative structure of secretion complexes, such as the Type 3 Secretion System (T3SS) from Gram-negative bacteria, using electron microscopy and cross-linking mass spectrometry data. This collaboration will both improve the understanding of the T3SS function and establish a state-of-the-art experimental and computational integrative structure determination pipelines at CSSB.
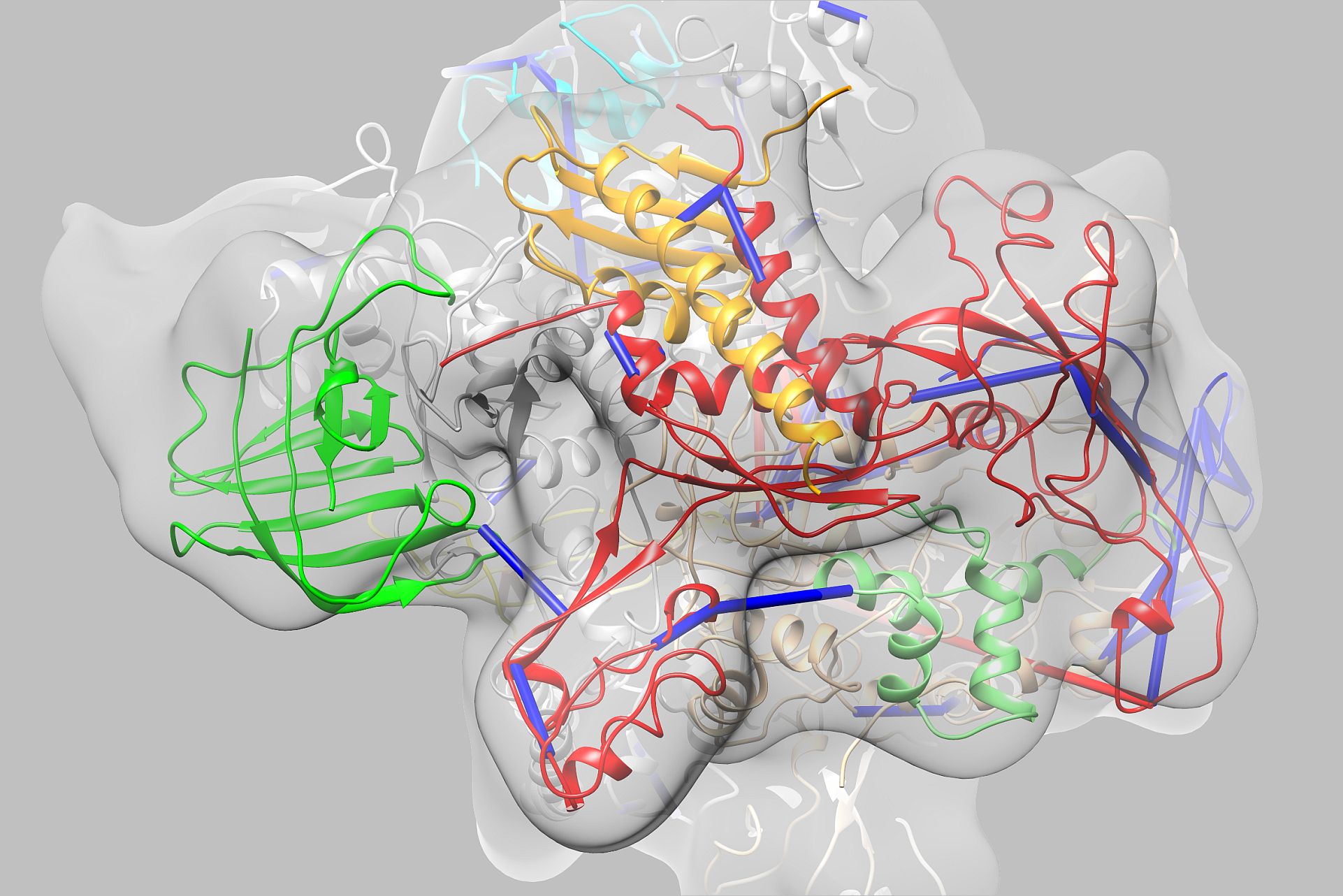
Research Groups: Michael Kolbe (HZI), Joerg Labahn (FZJ), and Susanne Häußler (HZI)
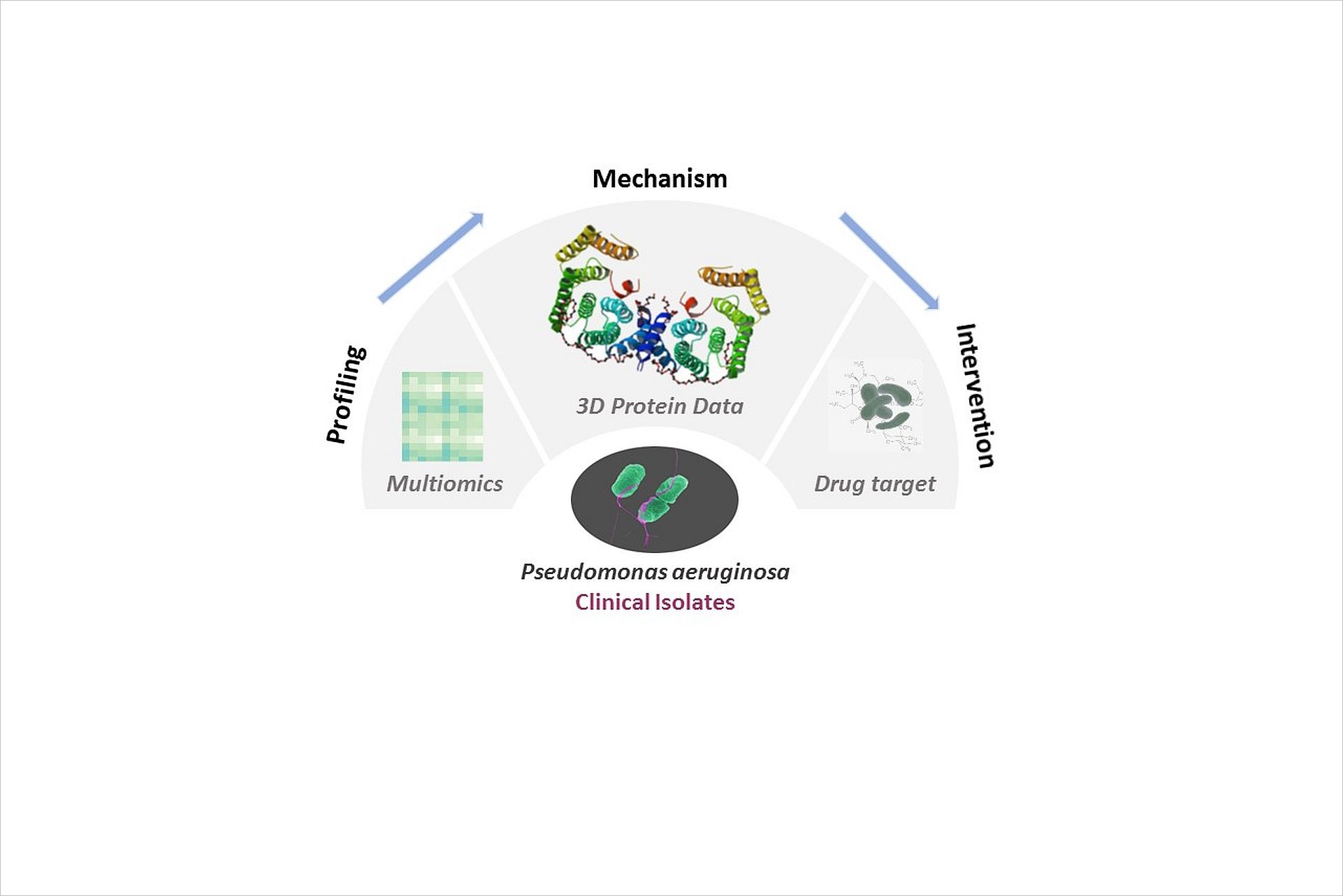
Global structure determination of virulence factors involved in nosocomial Pseudomonas aeruginosa infections as a model system for target discovery
Nosocomial infections represent a major concern in public health. One exceedingly problematic and opportunistic pathogen is Pseudomonas aeruginosa as it exhibits a remarkable ability to adapt to hostile environments and to develop multidrug-resistance. The aim of this project is the determination of three-dimensional structures and functional characterization of putative virulence proteins from uncharacterized open reading frames in P. aeruginosa. We will combine genomic, proteomic and transcriptional data of clinical isolates with microbiological, biophysical and structural biology methodologies to elucidate structure-function relationships of novel pathogenicity factors. This systems approach will set the bedrock for future structural programs searching for novel targets to treat infectious diseases at CSSB.
Research Groups: Kay Gruenewald (HPI/UHH), Tim Gilberger (BNITM/UHH), and Rainer Kaufmann (UHH)
Disassembly of the inner membrane complex of Plasmodium falciparum
The malaria parasite Plasmodium falciparum uses a complex molecular motor to drill itself into red blood cells. One critical part is a very unusual membrane structure (called inner membrane complex, IMC) that underlies the plasma membrane of the parasite. While the IMC plays a critical role for the host cell invasion, it completely disappears after this task is accomplished, most likely to enable the parasite to grow and proliferate within its host. To decipher the hitherto only rudimentarily understood process of IMC disassembly, a team of three CSSB research groups will apply an interdisciplinary approach using super-resolution microscopy, correlative light and electron microscopy (CLEM) and electron cryo tomography (cryoET) in combination with reverse genetics. This detailed investigation of IMC disassembly relies on CSSB’s core strength of combining outstanding technical infrastructure, operative expertise and innovation.



