Scientists Alter Membrane Proteins to Make Them Easier to Study
By making hydrophobic sections water-soluble, researchers hope to learn more about protein structures.
About 30 percent of the proteins encoded by the human genome are membrane proteins — proteins that span the cell membrane so they can facilitate communication between cells and their environment. Despite the prevalence of these proteins, scientists have had difficulty studying their structures and functions because the membrane-bound portions are very hydrophobic, so they cannot be dissolved in water. This makes it much harder to do structural analyses, such as X-ray crystallography and study molecular interactions.
In an advance that could make it easier to perform this type of structural study, and international team of researchers, including CSSB scientist Jörg Labahn, have developed a way to make these proteins water-soluble by swapping some of their hydrophobic amino acids for hydrophilic ones. This week, the study appears in the Proceedings of the National Academy of Sciences.
A simple code
Of the approximately 8,000 known membrane proteins found in human cells, scientists have discovered structures for about 50. They are widely viewed as very difficult to work with because once they are extracted from the cell membrane, they only maintain their structure if they are suspended in a detergent, which mimics the hydrophobic environment of the cell membrane. These detergents are expensive, and there is no universal detergent that works for all membrane proteins.
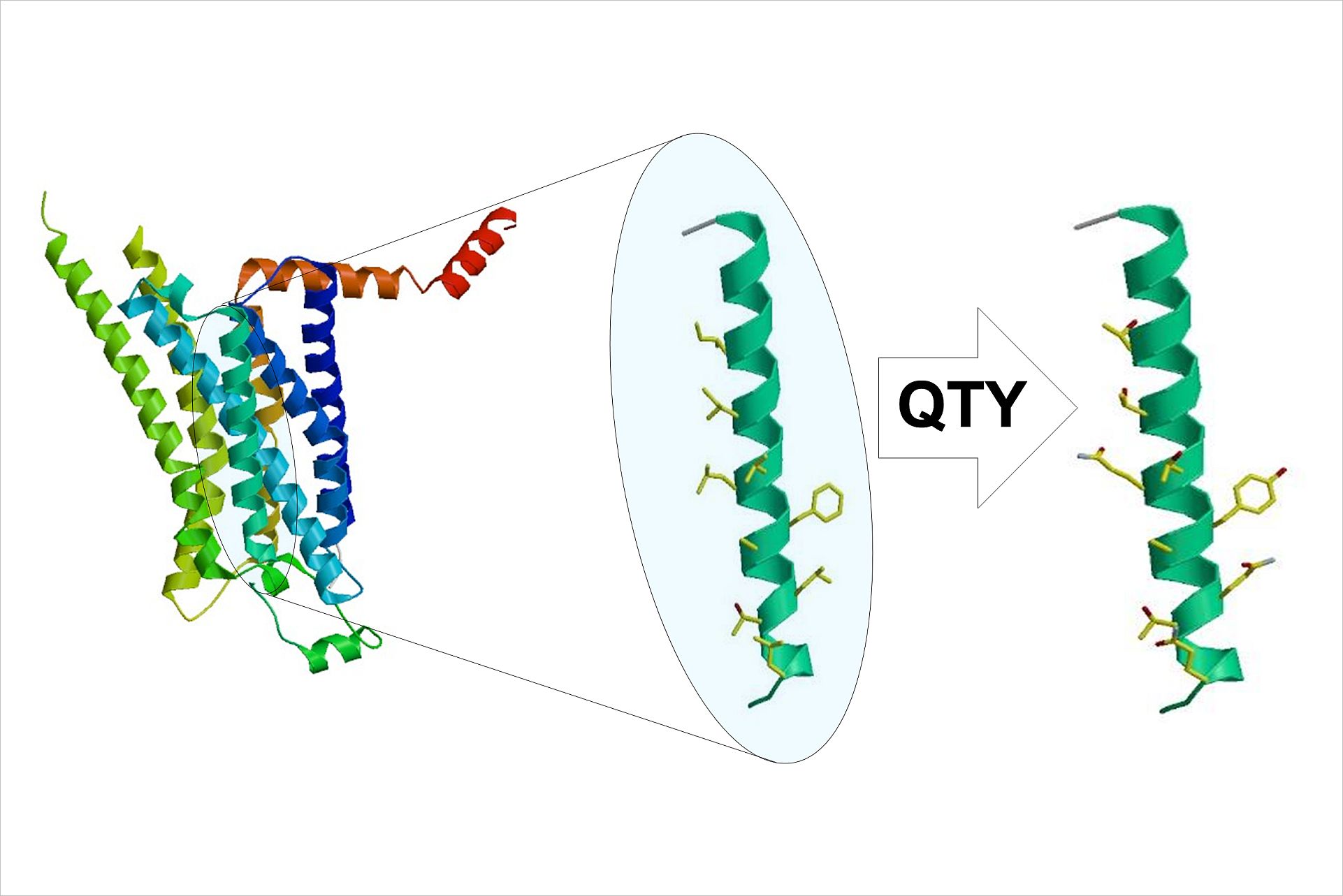
Shuguang Zhang, a scientist in the MIT Media Lab’s Center for Bits and Atoms, started working on whether protein structures called alpha helices, which make up the bulk of the membrane-embedded portion of proteins, could be switched from hydrophobic to hydrophilic. The key idea that allowed Zhang to develop the code is the fact that a handful of hydrophobic amino acids have very similar structures to some hydrophilic amino acids. These similarities allowed Zhang to come up with a code in which leucine is converted to glutamine, isoleucine and valine are converted to threonine, and phenylalanine is converted to tyrosine. The researchers call their code the QTY code, after the three letters that represent glutamine, threonine, and tyrosine, respectively.
Structural similarities
In this study, the researchers demonstrated their technique on four proteins that belong to a class of proteins known as G protein-coupled receptors. These proteins help cells to recognize molecules, such as hormones, or immune molecules, called chemokines, and trigger an appropriate response within the cell. “Although a number of scientists have been trying to find a way to ‘solubilize’ G protein-coupled receptors and other integral membrane proteins, until now their methods have not been of general use and often involved very complex computational methods that would not be widely applicable,” says Joel Sussman, a professor of structural biology at the Weizmann Institute of Science who was not involved in the research.
The researchers are still working towards obtaining the precise structures of these proteins using X-ray crystallography or nuclear magnetic resonance (NMR), but they performed some experiments that suggest the structures are similar. It may be possible to develop water-soluble versions of the proteins that bind to molecules normally expressed by cancer cells, which could be used to diagnose tumours or identify metastatic cancer cells in blood samples. “This decoy strategy could also be used for infectious diseases such as HIV that rely on GPCRS to infect cells,” explains Labahn. Water-soluble molecules could be created in which a membrane-bound receptor that viruses normally bind to is attached to part of an antibody. If these “decoy therapies” were injected into the body, viruses would bind to the receptors and then be cleared by the immune system, which would be activated by the antibody portion.
Original Publication:
QTY code enables design of detergent-free chemokine receptors that retain ligand-binding activities
Shuguang Zhang et al. Proceedings of the National Academy of Sciences (2018), doi.10.1073/pnas.1811031115
Original MIT Press Release:
http://news.mit.edu/2018/scientists-alter-membrane-proteins-make-them-easier-study-0827