A new method for identifying bacterial enzyme targets
CSSB Associate Member, Aymelt Itzen (UKE) and collaborators, Prof. Dr. Hartmut Schlüter from the UKE and Prof. Dr. Christian Hedberg from Sweden, have developed a new method that will systematically facilitate the identification of targets for a particular family of bacterial enzymes. Their chemical and biochemical method was recently published in Nature Chemistry.
Bacterial infections are the basis of numerous human diseases. Over the course of their developmental history, many pathogens have developed strategies to manipulate the infected host cell for their own benefit. Many bacteria release enzymes that can modify key components in human cells thus specifically influencing the cell’s function. The family of FIC proteins is particularly common in bacteria and in many cases causes the modification of host factors with an adenosine monophosphate group; a process is known as ampylation.
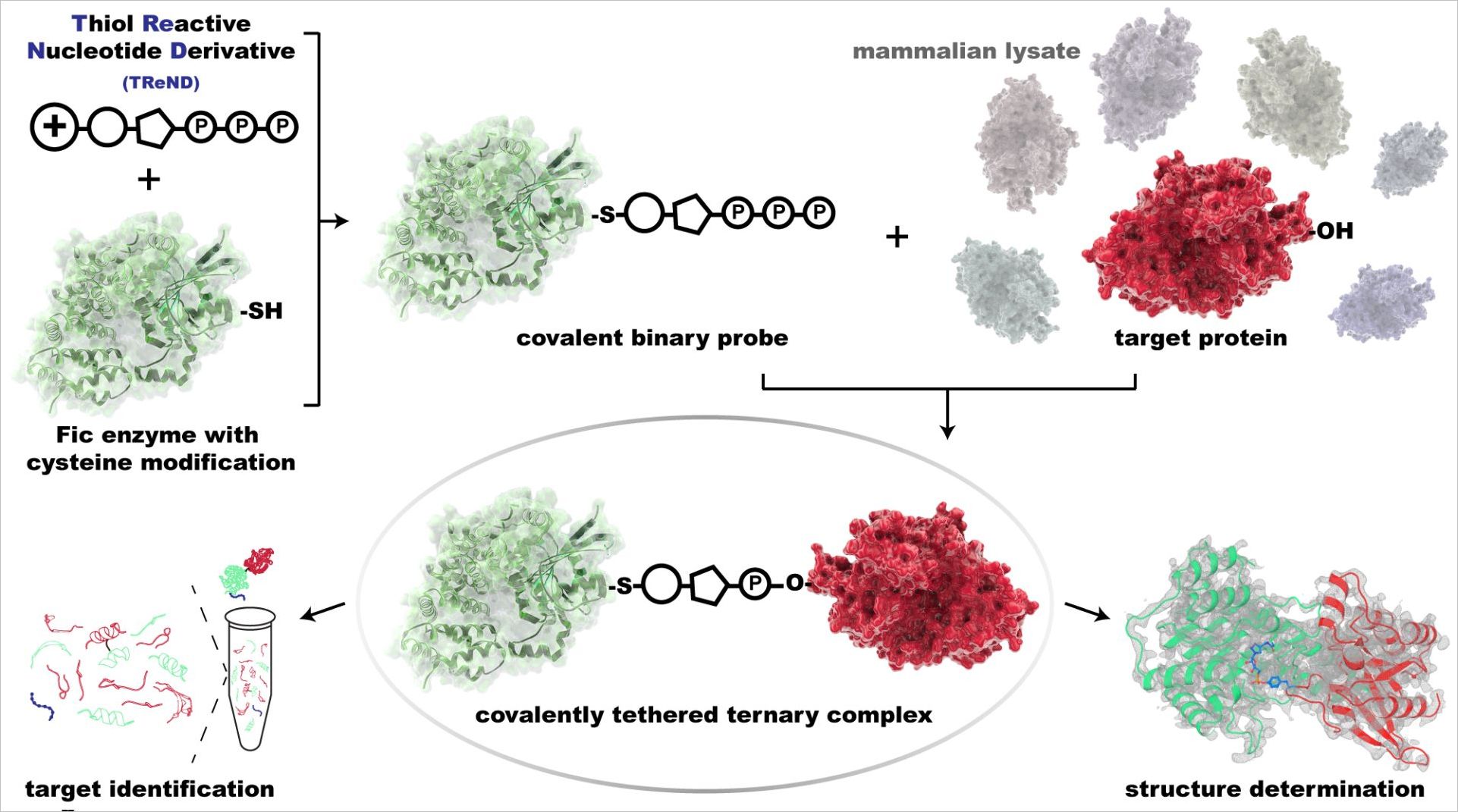
Despite existing knowledge of the FIC family and its discovery in bacteria, the human cellular targets cannot be easily predicted. Itzen and collaborators therefore developed a chemical and biochemical method to directly couple the targets of these bacterial manipulators with the enzyme and subsequently identify them. For this purpose, the chemistry of the adenosine monophosphates was chemically modified to form a permanent complex between the FIC enzyme and the human target protein. “With this approach we hope to be able to address FIC enzymes of relevant human pathogens in the near future and identify their targets in human cells,” explains Itzen “This could help lay the molecular foundations for a better understanding of bacterial diseases.”
CSSB scientist and associate members all seek to solve biological questions using a variety of complimentary methods and this project was no exception. Itzen emphasized that the combination of several approaches such as chemistry, biochemistry, structural biology and mass spectrometry was key to the success of this project.
Reference:
Gulen B, Rosselin M, Fauser J, Albers MF, Pett C, Krisp C, Pogenberg V, Schlüter H, Hedberg C, Itzen A. (2020) Identification of targets of AMPylating Fic enzymes by co-substrate-mediated covalent capture. Nat Chem;12(8):732-739. doi: 10.1038/s41557-020-0484-6.
Blog Post by Burak Gulen:
https://chemistrycommunity.nature.com/posts/co-substrate-mediated-covalent-enzyme-capture



