Understanding the Molecular Mechanisms of Salmonella Infection
To initiate infection, many Gram-negative bacteria inject bacterial proteins into human cells. The bacteria accomplish this by building a complex, syringe-like molecular machine known as the type III secretion system (T3SS). The Marlovits group (CSSB, UKE, DESY) together with CSSB’s light microscopy facility as well as collaborators from the University Medical Center Hamburg-Eppendorf (UKE) and the Hannover Medical School (MHH) have gained new structural insights into how proteins secreted by the TS33 subvert the host cell’s actin cytoskeleton during infection. Their findings have recently been published in the journal Scientific Advances.
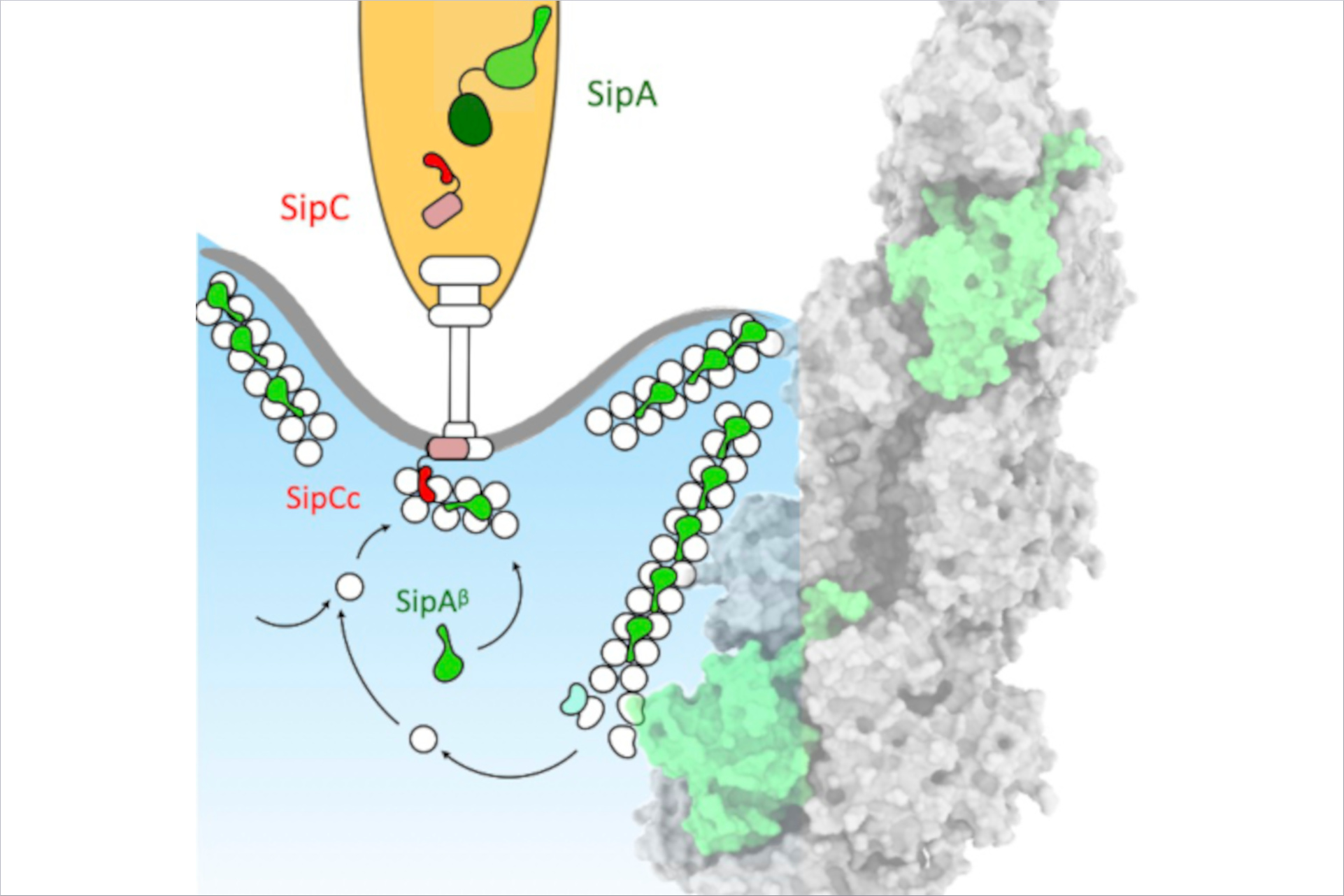
The shape of a cell is typically defined by its cytoskeleton. In the cytoskeleton lots of long protein filaments called actin filaments serve as a protein binding hubs which modify and interact with several other proteins in the cell. When Salmonella infects human cells its TS33 releases bacterial effectors some of which are actin-binding proteins (ABPs). The ABPs bind to the actin filaments in the human cell causing the cellular membrane to ruffle at the injection site. This local membrane ruffle enables the bacteria to be engulfed in the human host cell and propagate infection.
The researchers wanted to understand the specific interplay between two bacterial substrates, the translocator protein SipC and the effector protein SipA, which play a key role in the subversion of the actin cytoskeleton. Using the fluorescence microscopes available at CSSB’s Advanced Light and Fluorescence Microscopy (ALFM) Facility, the researchers examined the location of SipA and actin during Salmonella infection. “We discovered that SipA constantly modulates the actin network throughout the course of infection,” notes ALFM facility head Roland Thuenauer (CSSB, UHH, LIV).
To understand the underlying molecular mechanism of modulation of the cytoskeleton, the researchers used cryo-electron microscopy (cryo-EM), to generate high-resolution images of SipA bound to actin filaments. “We found that SipA binds to filamentous actin in a unique binding pattern and uses a specific binding site not utilized by the majority of actin-binding proteins,” notes Biao Yuan (CSSB, UKE) the paper’s first author. Together with collaborators from MHH and UKE, the researchers used total internal reflection fluoresce (TIRF) microscopy, to demonstrate that SipA not only helps to maintain a larger pool of actin filaments but also increases the stability of the filaments thus prolonging the membrane ruffles.
To initiate infection the needle part of the syringe-like T3SS latches on to the human host cell at the translocator proteins. "To our surprise, we found that the translocator protein SipC, which also contains an actin-binding-domain, cooperates in conjunction with SipA to efficiently decorate actin filaments. This cooperation is unanticipated because SipC remains bound to the host cell membrane, whereas SipA is localized in the cytoplasm,” notes Yuan.
“Overall understanding the interactions between these two substrates, helps to shed light on the coordinated sequence of infection events initiated by the T3SS in the host cell,” explains Thomas Marlovits the papers corresponding author “Given the unique nature of this binding mechanism, we plan to investigate the possibilities of inhibiting this process which could ultimately interfere with Salmonella invasion of host cells.”
Original Publication:
Yuan B, Scholz J, Wald J, Thuenauer R, Hennell James R, Ellenberg I, Windhorst S, Faix J, Marlovits TC (2023) Structural basis for subversion of host cell actin cytoskeleton during Salmonella infection. Sci Adv. 9(49):eadj5777. doi: 10.1126/sciadv.adj5777.



