New Family of Bacterial Enzymes
The Sondermann Group (DESY, CAU) at CSSB along with international collaborators have discovered a novel family of bacterial enzymes that function as dedicated deoxydinucleases and may act as part of bacterial immunity. A description of this new enzyme family, which have been termed “diDNases,” has been published in Nucleic Acids Research where it received a “breakthrough study” designation.
Within a cell, chemical reactions are constantly occurring to allow for the synthesis and degradation of the nucleic acids, DNA and RNA. These chemical reactions are driven by enzymes. During RNA degradation various classes of enzymes work together to break down polymers into smaller molecules. The ultimate step in this process, the degradation of dinucleotides to mononulceotides, requires specialized enzymes, so-called dinucleases such as oligoribonuclease or nano-RNase C.
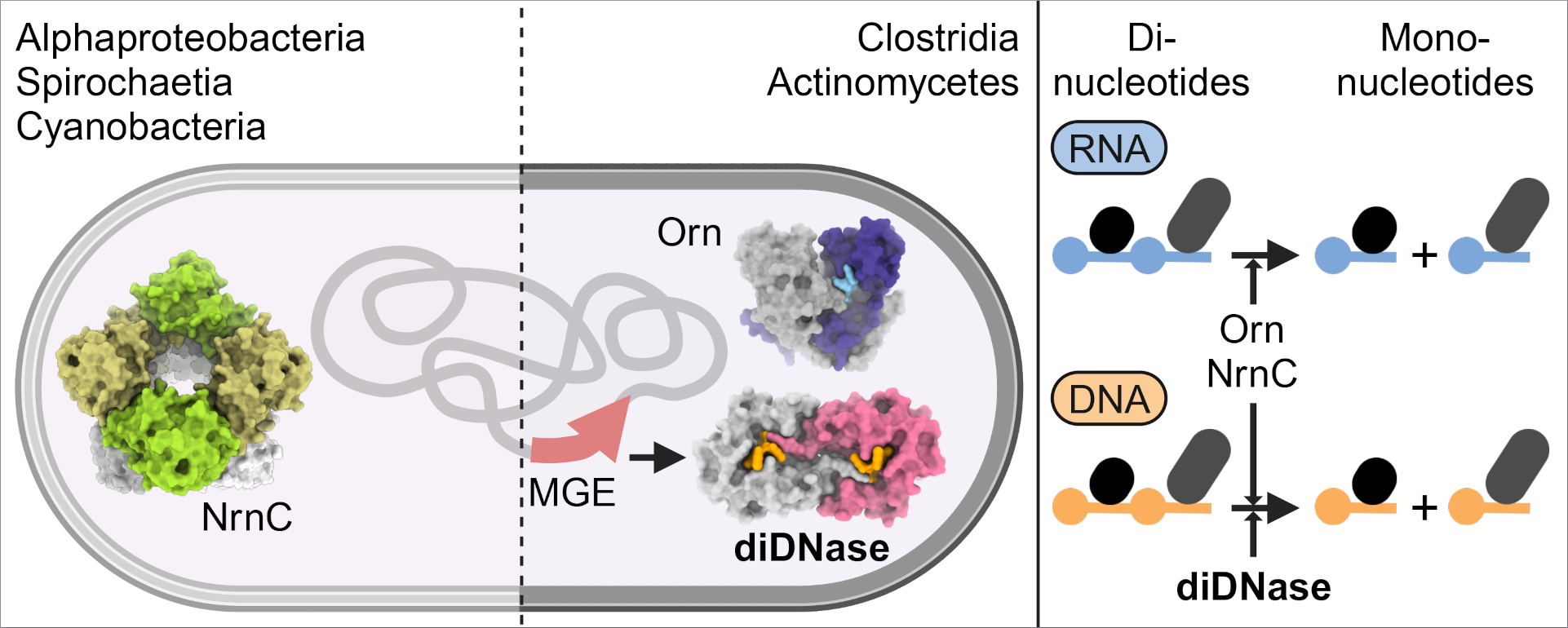
Previous work from the collaborative team has been shown that the active sites of these specialized enzymes are optimized for substrates that are two nucleotides long and do not discriminate between RNA and DNA. In their new study, using biophysical analysis performed at CSSB’s SPC Facility run by EMBL in combination with x-ray crystallography experiments performed at DESY’s PETRA III beamline P11, the Sondermann group identified a novel enzyme subfamily whose members function as dedicated deoxydinucleases.
“These newly identified diDNases specifically hydrolyze single-stranded DNA dinucleotides in a sequence-independent manner,” explains the study’s first author Sofia Mortensen “In fact, our crystal structures of enzyme-substrate complexes reveal that diDNases’ specificity for DNA stems from a combination of conserved structural elements.” The structural insights revealed a mechanistic basis for DNA specificity, which excludes the dinucleotides from RNA as possible substrates. “diDNases stand out for their lack of promiscuity in substrate preference,” notes Mortensen.
In subsequent, cell-based experiments, the team could show that diDNases failed to fully replace the more promiscuous dinucleases. “We can therefore conclude that diDNases have a distinct cellular role,” notes Holger Sondermann the study’s corresponding author “their association with phage-defence systems and the fact that the genes encoding diDNases are located in mobile genetic elements of Gram-positive bacteria suggest that diDNases play a role in bacterial immunity.”
Only 2-3% of the papers accepted by Nucleic Acids Research are designated as breakthrough, manuscripts that answer long-standing questions in the field or motivate new research directions. The study by Mortensen et al. falls into the latter category. While RNA-based dinucleotides are well-known secondary metabolites and signaling molecules, this study implicates DNA dinucleotides in cellular processes, broadening the scope of biologically relevant dinucleotides.
“We are honored to have received the breakthrough designation,” notes Sondermann “this underscores the importance of fundamental, experimental research. We are looking forward to conducting further research into the function of diDNases and discovering more about their involvement in bacterial immunity.”
The investigators and work in this study was supported by the National Institutes of Health (USA) and the European Union’s Horizon 2020 research and innovation program. It is based on a close collaboration involving the groups of Drs. Vincent T. Lee and Wade C. Winkler at the University of Maryland, USA.
Oringinal Publication:
Mortensen S, Kuncová S, Lormand JD, Myers TM, Kim S-K, Lee VT, Winkler WC, Sondermann H, Structural and bioinformatics analyses identify deoxydinucleotide-specific nucleases and their association with genomic islands in gram-positive bacteria, Nucleic Acids Research, Volume 53, Issue 1, 13 January 2025, gkae1235, https://doi.org/10.1093/nar/gkae1235



