Research Projects
RESEARCH AT CSSB
Malaria, caused by protozoan parasites of the genus Plasmodium continues to be a major global health burden, with an estimated 450,000 deaths annually and half of the world’s population at risk of infection. There is still no reliable vaccine available and resistance against frontline anti-malaria drugs is emerging. Thus, new drugs and intervention strategies are urgently needed, in order to fight the parasite and get closer to the goal of malaria eradication. The sexual gametocyte stages, which are taken up from the blood by Anopheles mosquitoes, are of special interest in this context. Being the only developmental stage of the parasite that is transmissible between human and mosquito hosts, they provide a prime target for new transmission-blocking intervention strategies. However, despite their high relevance for such strategies, the molecular mechanisms driving the switch from asexual replication to sexual development of malaria parasites remain poorly understood.
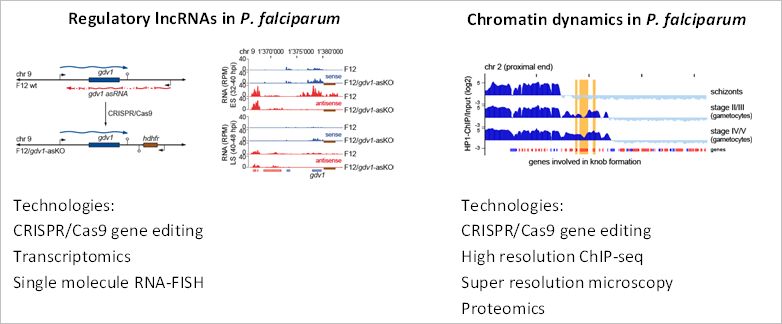
During asexual replication in human red blood cells, P. falciparum uses the conserved mechanism of heterochromatin protein 1 (HP1)-mediated gene silencing to epigenetically control sexual commitment and prevent the development of gametocytes. Changes in environmental conditions can lead to a remodelling of chromatin structure and the disassembly of heterochromatin across specific regions of the genome. This leads to a cell cycle switch and the activation of gametocyte development. Recent research from us and other laboratories identified key molecular factors involved in this process. In addition, we were able to show that regulatory long non-coding (lnc) RNAs play a part in this epigenetic network.
However, how environmental triggers set off this lncRNA-mediated regulation, how the involved factors are targeted to specific loci in the genome and what molecular machinery governs the establishment of active and inactive chromatin structures across the nucleus remains unknown. Our lab is focused on answering these questions, by investigating the formation of active and inactive chromatin structures and the molecular machinery governing these highly dynamic processes in P. falciparum. In addition, we are focused on uncovering the functions of lncRNAs in the regulation of these processes.
Ultimately, we believe that this will enable us to develop a broader understanding of the connection between environmental signals, changes of chromatin architecture and the epigenetic networks, which control sexual commitment and gametocyte development in malaria parasites.
KEY RESEARCH QUESTIONS
- How is the expression of lncRNAs controlled?
- What roles do lncRNAs play in the epigenetic control of gene expression and cell cycle control?
- What is the molecular machinery governing chromatin dynamics in P. falciparum?