Research Projects
PREVIOUS AND CURRENT RESEARCH
Understanding the molecular processes of life requires an understanding of proteins: Almost every biological process involves these biomolecules. It is therefore not surprising that detailed studies of proteins at the molecular level have provided fundamental insights into human biochemistry and cell biology.
Our group studies the structures, regulation, and biochemistry of proteins and protein complexes. In particular, the contribution of posttranslational modifications (PTMs) to the regulation of protein activity is a central research topic in our laboratory.
In this respect, it is interesting to consider the influence of pathogenic bacteria on the function and activity of human proteins: During evolution, bacterial pathogens have developed sophisticated strategies to escape their elimination by the human immune system. During the infection process, several bacterial pathogens release a multitude of effector proteins which interfere with essential intracellular mechanisms in the host cell . The study of the molecular basis of such manipulation processes enables us to identify and describe strategies of pathogens.
Our goal is to understand the underlying mechanisms of manipulation of human proteins using selected bacterial factors in molecular detail. Therefore, we isolate the desired bacterial proteins in high purity and study their biochemical and functional properties using biophysical and structural biology methods.
PTMs in the context of disease and infection
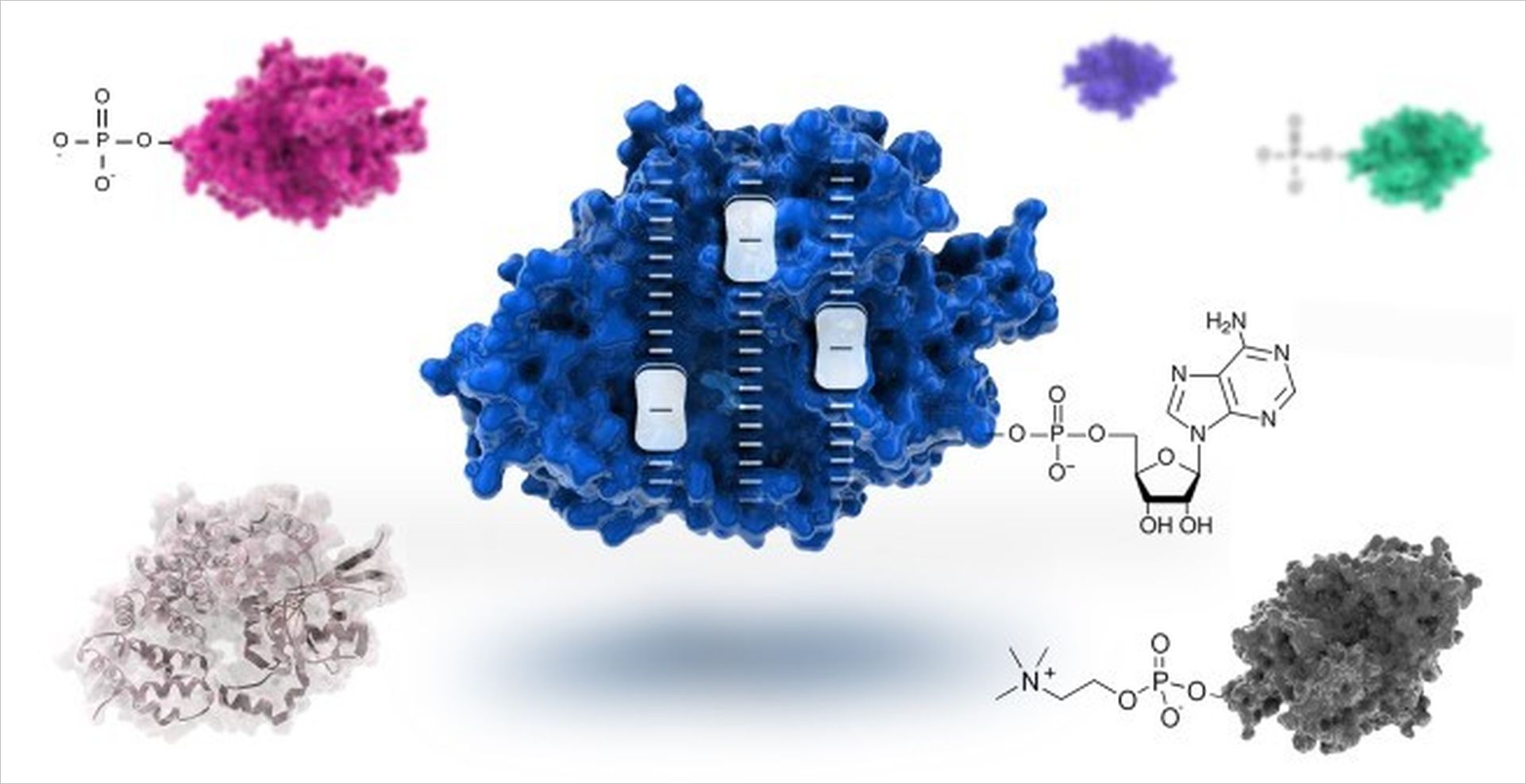
A current focus of our work is the investigation of post-translational modifications (PTMs) that occur in the context of bacterial pathogens. PTMs are chemical changes in proteins caused by enzymes. These transformations significantly influence the activity and functionality of the modified proteins. Therefore, many pathogens release enzymes that selectively and specifically modify central factors of human cells to give the pathogen an advantage.
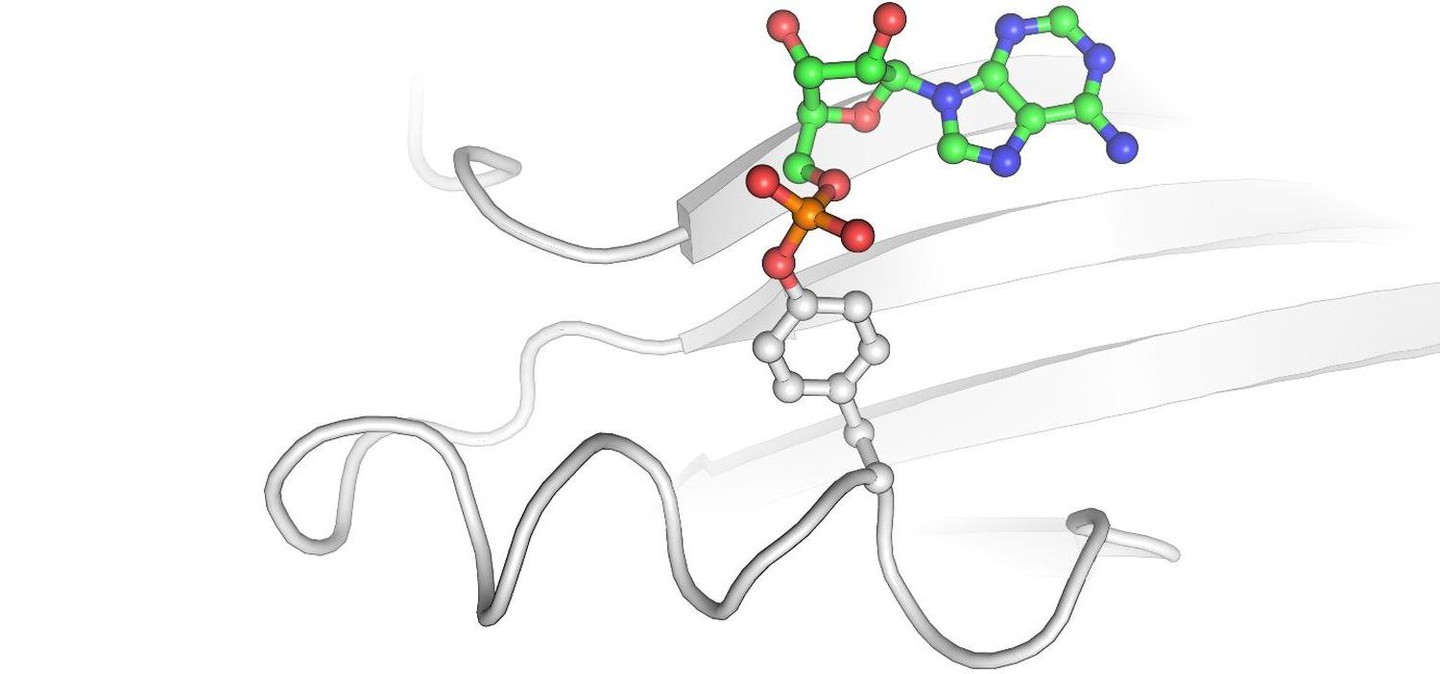
Of particular interest for us among the post-translational modifications is the so-called AMPylation of human proteins. Many bacterial pathogens inject enzymes into host cells that use the generally available adenosine triphosphate (ATP) to link target proteins with an adenosine monophosphate (AMP). We now know that the AMP-transmitting enzymes are present in many bacterial pathogens, but their target proteins cannot be predicted.
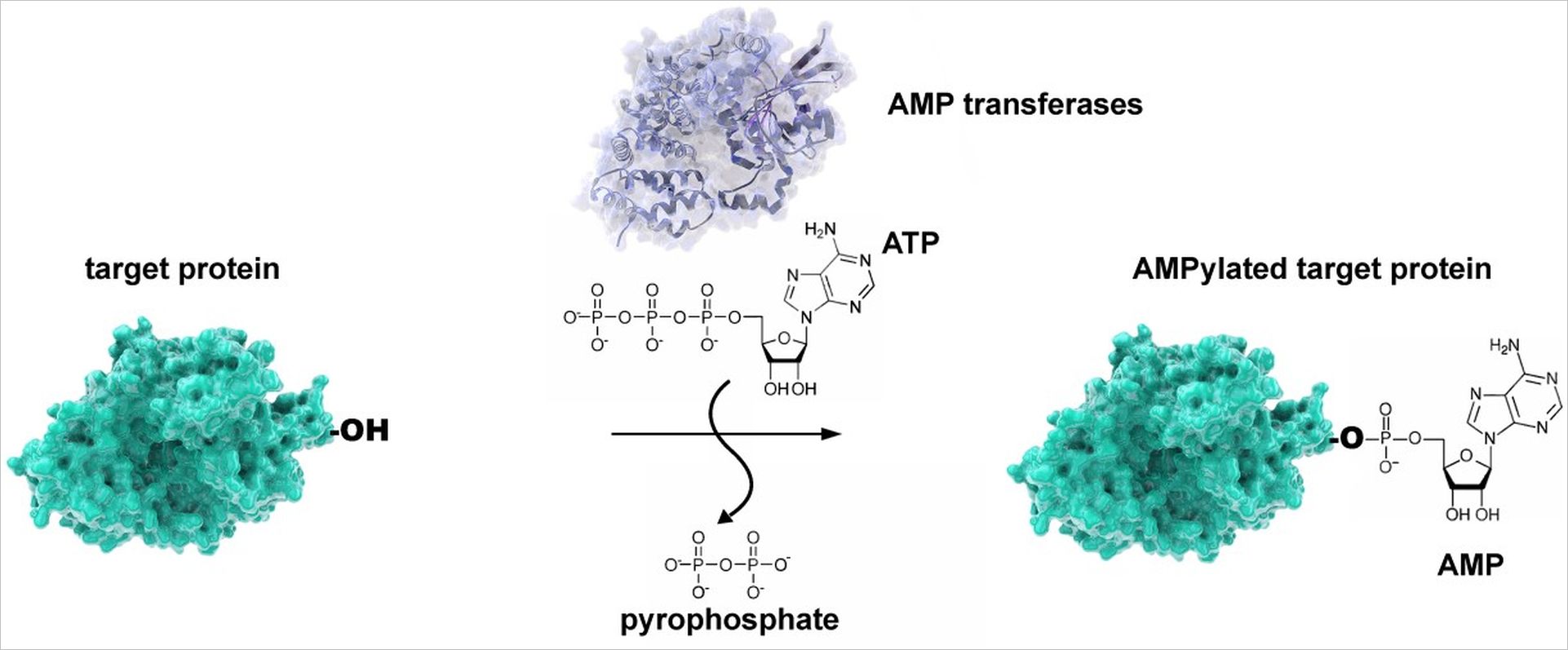
Therefore, a central topic of our work is the development of methods to identify AMP-modified proteins. For this purpose, we use a spectrum of biochemical, chemical, mass spectrometric and immunological methods, which allow the targeted enrichment and analysis of AMPylated molecules. But also, other PTMs (phosphocholination, phosphorylation, proteolysis) are subject of our research.
In addition, we want to understand the biochemical, functional, and structural consequences of PTMs in the context of bacterial infections in molecular detail. The introduction and analysis of PTMs (e.g., AMPylations) are technically challenging and require extensive knowledge of the biochemistry and function of the respective proteins and enzymes. A core expertise of our group is therefore the generation of proteins, the introduction of PTMs, and the comprehensive characterization of these molecules. Our research approach enables us to identify targets of bacteria and to study their possible cellular consequences.
FUTURE PROJECTS AND GOALS
- Understanding the molecular and functional consequences of bacterially caused PTMs.
- Examining the atomic structures of PTM-transferring enzymes in complex with targets.
- Elucidating crosstalk of PTMs during infections by bacterial pathogens.
- Developing methods for systematic identification of selected PTMs during infection.
SELECTED PUBLICATIONS
Gulen, B., Itzen, A. (2021) Revisiting AMPylation through the lens of Fic enzymes. Trends Microbiol.https://pubmed.ncbi.nlm.nih.gov/34531089/
Ernst, S., Ecker, F., Kaspers, M.S., Ochtrop, P., Hedberg, C., Groll, M. & Itzen, A. (2020) Legionella effector AnkX displaces the switch II region for Rab1b phosphocholination. Sci. Adv. 6, eaaz8041. https://www.ncbi.nlm.nih.gov/pubmed/32440549
Barthelmes, K., Ramcke, E., Kang, H.S., Sattler, M. & Itzen, A. (2020) Conformational control of small GTPases by AMPylation. Proc. Natl. Acad. Sci. U.S.A. 117, 5772-5781. https://www.ncbi.nlm.nih.gov/pubmed/32123090
Du, J., Wrisberg, M.V., Gulen, B., Stahl, M., Pett, C., Hedberg, C., Lang, K., Schneider, S. & Itzen, A. (2021) Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1. Nat. Commun. 12, 460. https://www.ncbi.nlm.nih.gov/pubmed/33469029
Wachtel, R., Brauning, B., Mader, S.L., Ecker, F., Kaila, V.R.I., Groll, M. & Itzen, A. (2018) The protease GtgE from Salmonella exclusively targets inactive Rab GTPases. Nat. Commun. 9, 44. https://www.ncbi.nlm.nih.gov/pubmed/29298974
Hopfner, D., Fauser, J., Kaspers, M.S., Pett, C., Hedberg, C. & Itzen, A. (2020) Monoclonal Anti-AMP Antibodies Are Sensitive and Valuable Tools for Detecting Patterns of AMPylation. iScience 23, 101800. https://www.ncbi.nlm.nih.gov/pubmed/33299971
Gulen, B., Rosselin, M., Fauser, J., Albers, M.F., Pett, C., Krisp, C., Pogenberg, V., Schluter, H., Hedberg, C. & Itzen, A. (2020) Identification of targets of AMPylating Fic enzymes by co-substrate-mediated covalent capture. Nat. Chem. 12, 732-739. https://www.ncbi.nlm.nih.gov/pubmed/32632184
Fauser, J., Gulen, B., Pogenberg, V., Pett, C., Pourjafar-Dehkordi, D., Krisp, C., Hopfner, D., Konig, G., Schluter, H., Feige, M.J., Zacharias, M., Hedberg, C. & Itzen, A. (2021) Specificity of AMPylation of the human chaperone BiP is mediated by TPR motifs of FICD. Nat. Commun. 12, 2426. https://www.ncbi.nlm.nih.gov/pubmed/33893288