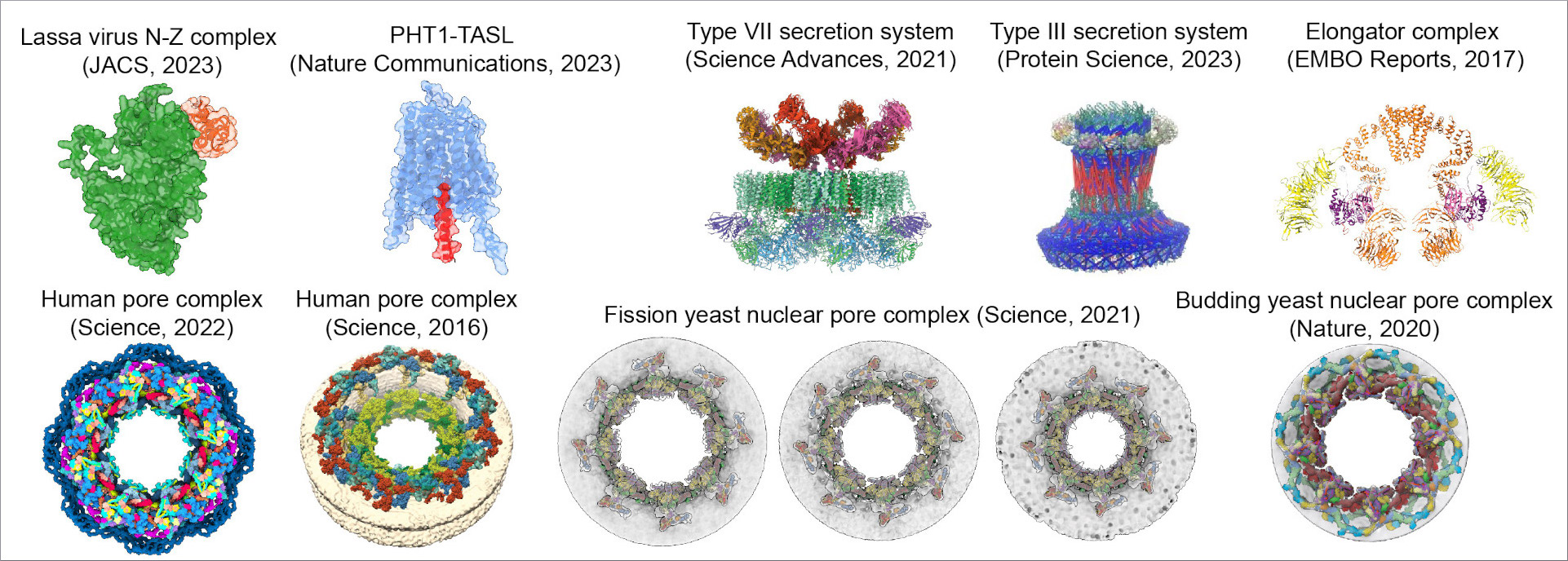
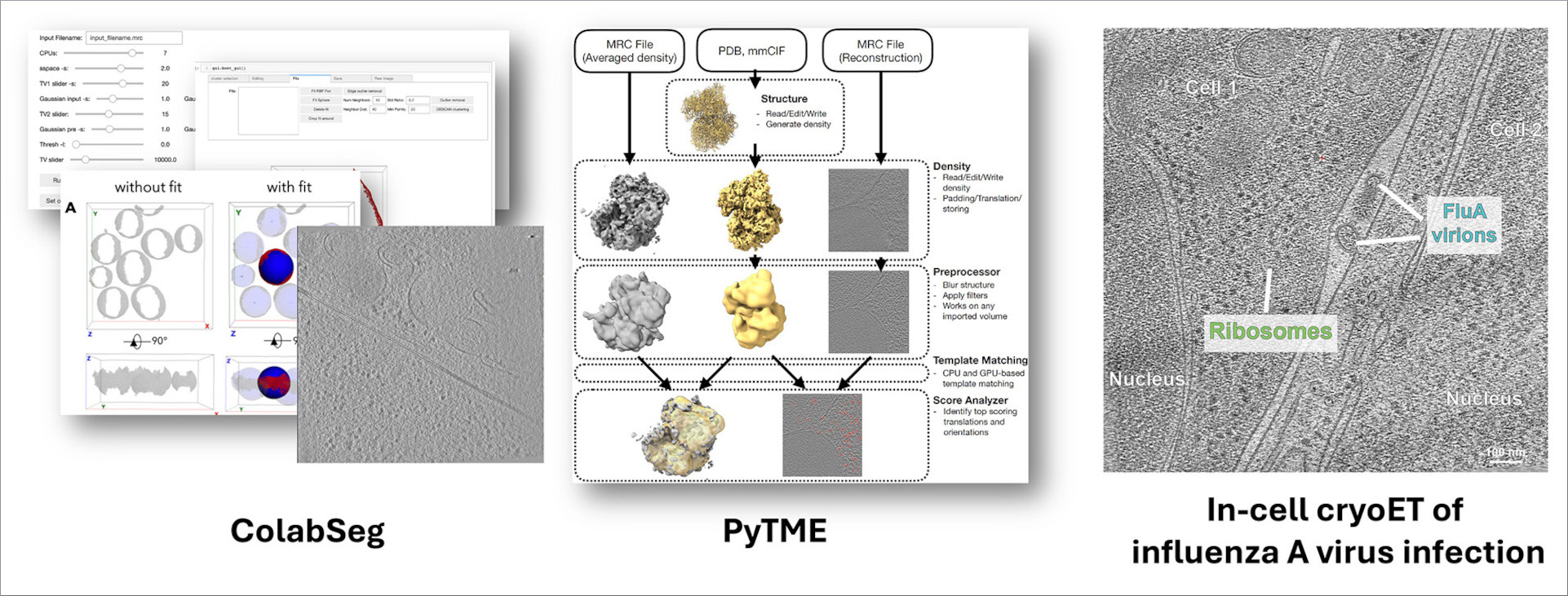
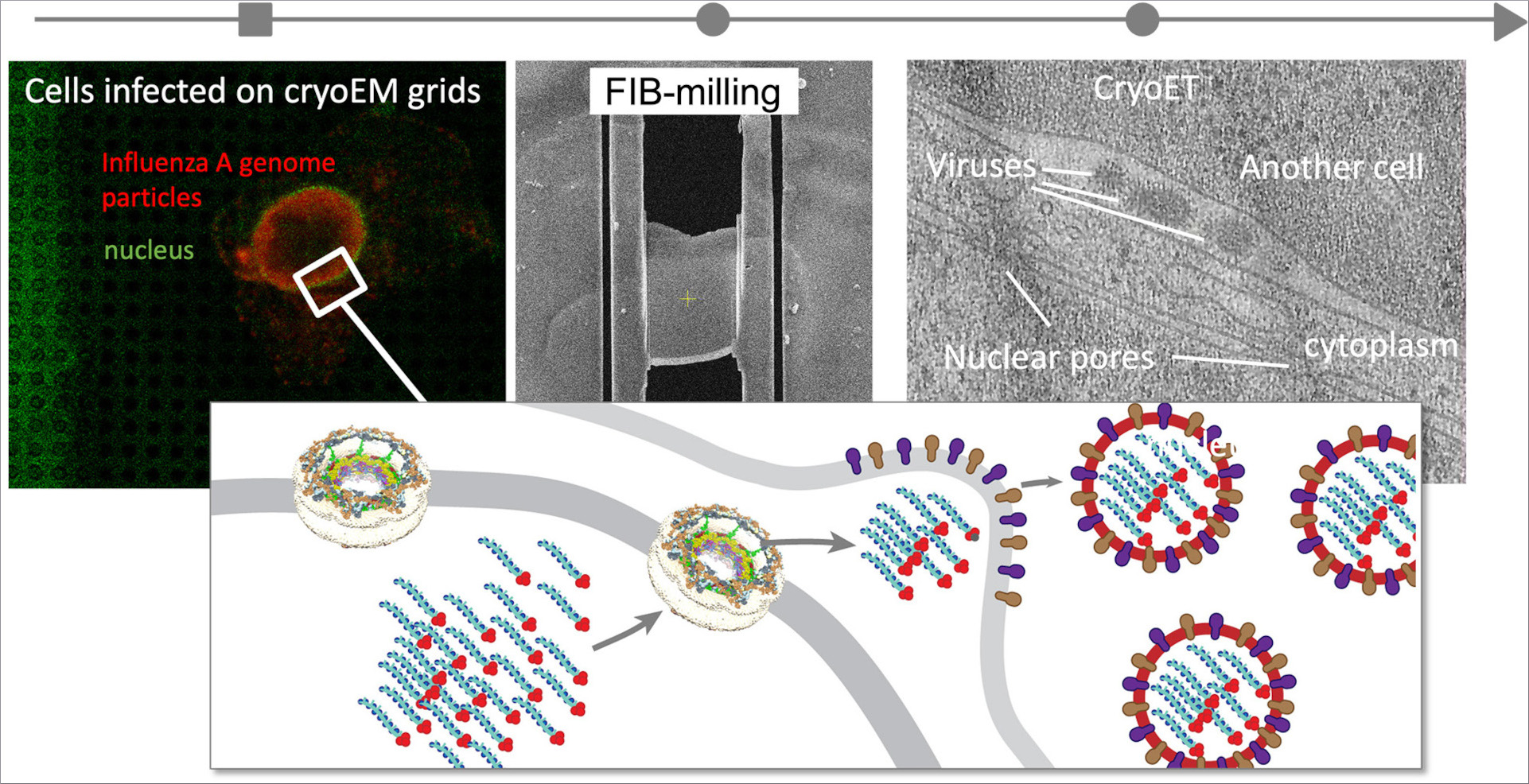
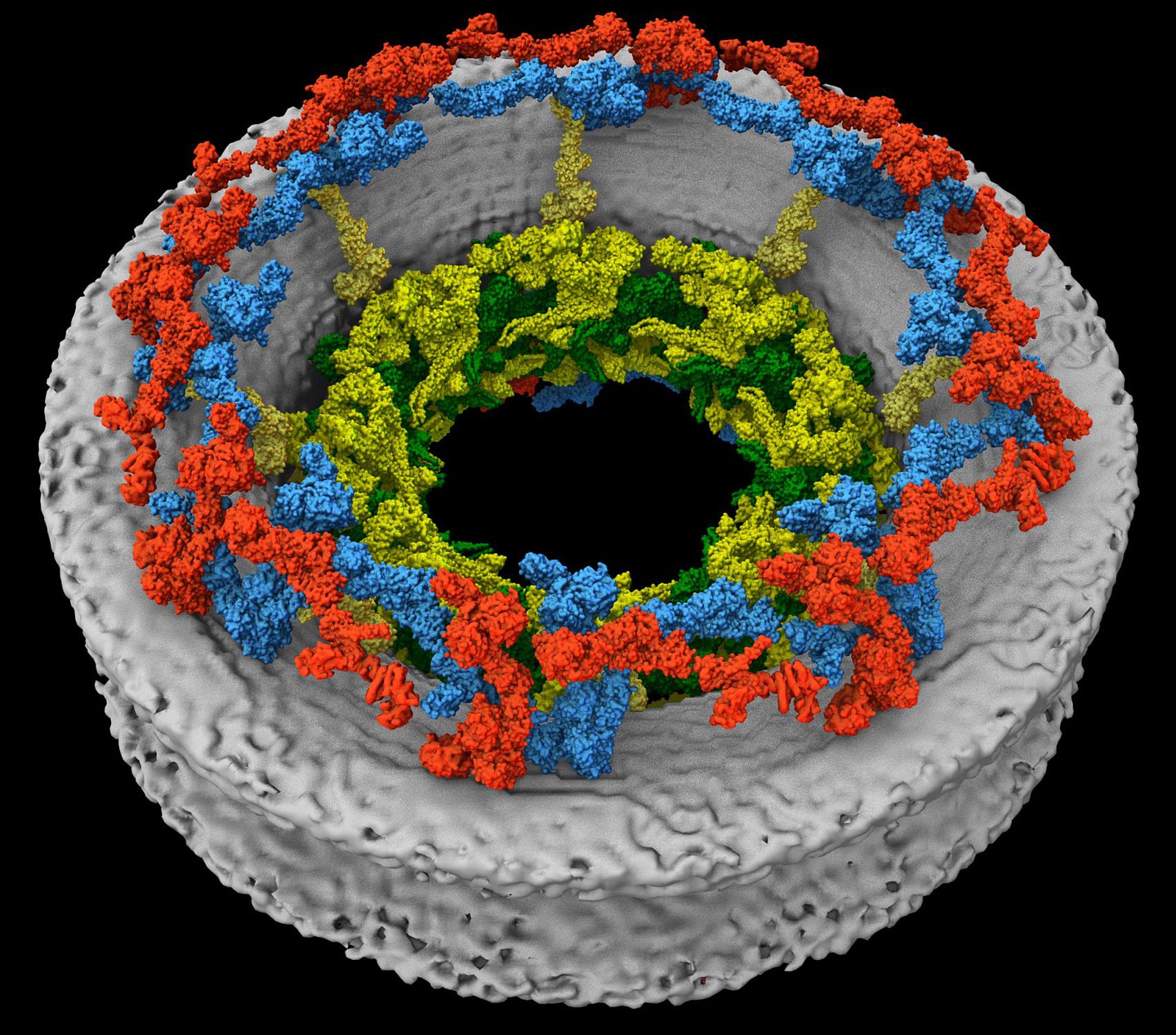
Kotova I, Mühlberg L, Gilep K, Yu D, Ziemianowicz D, Stanelle-Bertram S, Beck S, Baeg K, Duss O, Gabriel G, Liu F, Bogdanow B, Kosinski J (2025) Snapshot of in-cell protein contact sites reveals new host factors and hijacking of paraspeckles during influenza A virus infection. bioRxiv 642134; doi: 10.1101/2025.03.09.642134
Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salomé PA, Merchant SS, Plitzko JM, Baumeister W, Engel BD, Beck M (2018) In situ architecture of the algal nuclear pore complex. Nat Commun 9: 2361 doi: 10.1038/s41467-018-04739-y
Mathieson T, Franken H, Kosinski J, Kurzawa N, Zinn N, Sweetman G, Poeckel D, Ratnu VS, Schramm M, Becher I, Steidel M, Noh KM, Bergamini G, Beck M, Bantscheff M, Savitski MM (2018) Systematic analysis of protein turnover in primary cells. Nat Commun 9: 689 doi: 10.1038/s41467-018-03106-1
Beck M, Mosalaganti S, Kosinski J (2018) From the resolution revolution to evolution: structural insights into the evolutionary relationships between vesicle coats and the nuclear pore. Curr Opin Struct Biol 52: 32-40 doi: 10.1016/j.sbi.2018.07.012
2017
Teimer R, Kosinski J, von Appen A, Beck M, Hurt E (2017) A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat Commun 8: 1107 doi: 10.1038/s41467-017-01160-9
Sadian Y, Tafur L, Kosinski J, Jakobi AJ, Wetzel R, Buczak K, Hagen WJ, Beck M, Sachse C, Müller CW (2017) Structural insights into transcription initiation by yeast RNA polymerase I. EMBO J 36: 2698-2709 doi: 10.15252/embj.201796958
Dauden MI, Kosinski J, Kolaj-Robin O, Desfosses A, Ori A, Faux C, Hoffmann NA, Onuma OF, Breunig KD, Beck M, Sachse C, Séraphin B, Glatt S, Müller CW (2017) Architecture of the yeast Elongator complex. EMBO Rep 18: 264-279 doi: 10.15252/embr.201643353
2016
Thierry E, Guilligay D, Kosinski J, Bock T, Gaudon S, Round A, Pflug A, Hengrung N, El Omari K, Baudin F, Hart DJ, Beck M, Cusack S (2016) Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of PB2 Domains. Mol Cell 61: 125-137 doi: 10.1016/j.molcel.2015.11.016
Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, Hurt E, Beck M (2016) Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352: 363-365 doi: 10.1126/science.aaf0643
Hoffmann NA, Sadian Y, Tafur L, Kosinski J, Müller CW (2016) Specialization versus conservation: How Pol I and Pol III use the conserved architecture of the pre-initiation complex for specialized transcription. Transcription 7: 127-132 doi: 10.1080/21541264.2016.1203628
Ferber M, Kosinski J, Ori A, Rashid UJ, Moreno-Morcillo M, Simon B, Bouvier G, Batista PR, Müller CW, Beck M, Nilges M (2016) Automated structure modeling of large protein assemblies using crosslinks as distance restraints. Nat Methods 13: 515-520 doi: 10.1038/nmeth.3838
Bertipaglia C, Schneider S, Jakobi AJ, Tarafder AK, Bykov YS, Picco A, Kukulski W, Kosinski J, Hagen WJ, Ravichandran AC, Wilmanns M, Kaksonen M, Briggs JA, Sachse C (2016) Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle. EMBO Rep 17: 1044-1060 doi: 10.15252/embr.201541960
2015
von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, Buczak K, Mosalaganti S, Hagen W, Andres-Pons A, Lemke EA, Bork P, Antonin W, Glavy JS, Bui KH, Beck M (2015) In situ structural analysis of the human nuclear pore complex. Nature 526: 140-143 doi: 10.1038/nature15381
Kosinski J, von Appen A, Ori A, Karius K, Müller CW, Beck M (2015) Xlink Analyzer: software for analysis and visualization of cross-linking data in the context of three-dimensional structures. J Struct Biol 189: 177-183 doi: 10.1016/j.jsb.2015.01.014
Hoffmann NA, Jakobi AJ, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen WJ, Sachse C, Müller CW (2015) Molecular structures of unbound and transcribing RNA polymerase III. Nature 528: 231-236 doi: 10.1038/nature16143
2013
Kosinski J*, Barbato A*, Tramontano A (2013) MODexplorer: an integrated tool for exploring protein sequence, structure and function relationships. Bioinformatics 29: 953–4.
2012
Barbato A, Benkert P, Schwede T, Tramontano A, Kosinski J, (2012) Improving your target-template alignment with MODalign. Bioinformatics 28: 1038–9.
2011
Milanowska K, Krwawicz J, Papaj G, Kosinski J, Poleszak K, Lesiak J, Osinska E, Rother K, Bujnicki JM (2011) REPAIRtoire--a database of DNA repair pathways. Nucleic Acids Res. 39: D788–92.
2010
Kosinski J, Hinrichsen I, Bujnicki JM, Friedhoff P, Plotz G (2010) Identification of Lynch syndrome mutations in the MLH1-PMS2 interface that disturb dimerization and mismatch repair. Hum. Mutat. 31: 975–82.
Pukáncsik M, Békési A, Klement E, Hunyadi-Gulyás E, Medzihradszky KF, Kosinski J, Bujnicki JM, Alfonso C, Rivas G, Vértessy BG (2010) Physiological truncation and domain organization of a novel uracil-DNA-degrading factor. FEBS J. 277: 1245–59.
2008
Kosinski J*, Plotz G*, Guarné A, Bujnicki JM, Friedhoff P (2008) The PMS2 subunit of human MutLalpha contains a metal ion binding domain of the iron-dependent repressor protein family. J. Mol. Biol. 382: 610–27.
2007
Chovancová E, Kosinski J, Bujnicki JM, Damborský J (2007) Phylogenetic analysis of haloalkane dehalogenases. Proteins 67: 305–16.
Kosinski J, Kubareva E, Bujnicki JM (2007) A model of restriction endonuclease MvaI in complex with DNA: a template for interpretation of experimental data and a guide for specificity engineering. Proteins 68: 324–36.
Liu Y, Li Z, Lin Q, Kosinski J, Seetharaman J, Bujnicki JM, Sivaraman J, Hew C-L (2007) Structure and evolutionary origin of Ca(2+)-dependent herring type II antifreeze protein. PLoS One 2: e548.
Miyazono K, Watanabe M, Kosinski J, Ishikawa K, Kamo M, Sawasaki T, Nagata K, Bujnicki JM, Endo Y, Tanokura M, Kobayashi I (2007) Novel protein fold discovered in the PabI family of restriction enzymes. Nucleic Acids Res. 35: 1908–18.
2006
Ahrends R, Kosinski J, Kirsch D, Manelyte L, Giron-Monzon L, Hummerich L, Schulz O, Spengler B, Friedhoff P (2006) Identifying an interaction site between MutH and the C-terminal domain of MutL by crosslinking, affinity purification, chemical coding and mass spectrometry. Nucleic Acids Res. 34: 3169–80.
Skowronek KJ, Kosinski J, Bujnicki JM (2006) Theoretical model of restriction endonuclease HpaI in complex with DNA, predicted by fold recognition and validated by site-directed mutagenesis. Proteins 63: 1059–68.
Zegers I, Gigot D, van Vliet F, Tricot C, Aymerich S, Bujnicki JM, Kosinski J, Droogmans L (2006) Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase. Nucleic Acids Res. 34: 1925–34.
2005
Armalyte E, Bujnicki JM, Giedriene J, Gasiunas G, Kosiński J, Lubys A (2005) Mva1269I: a monomeric type IIS restriction endonuclease from Micrococcus varians with two EcoRI- and FokI-like catalytic domains. J. Biol. Chem. 280: 41584–94.
Kosinski J, Feder M, Bujnicki JM (2005) The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics 6: 172.
Kosinski J, Gajda MJ, Cymerman IA, Kurowski MA, Pawlowski M, Boniecki M, Obarska A, Papaj G, Sroczynska-Obuchowicz P, Tkaczuk KL, Sniezynska P, Sasin JM, Augustyn A, Bujnicki JM, Feder M (2005) Frankenstein becomes a cyborg: the automatic recombination and realignment of fold recognition models in CASP6. Proteins 61 Suppl 7: 106–13.
Kosinski J, Steindorf I, Bujnicki JM, Giron-Monzon L, Friedhoff P (2005) Analysis of the quaternary structure of the MutL C-terminal domain. J. Mol. Biol. 351: 895–909.
2004
Ye X, O’Neil PK, Foster AN, Gajda MJ, Kosinski J, Kurowski MA, Bujnicki JM, Friedman AM, Bailey-Kellogg C (2004) Probabilistic cross-link analysis and experiment planning for high-throughput elucidation of protein structure. Protein Sci. 13: 3298–313.
2003
Kosinski J, Cymerman IA, Feder M, Kurowski MA, Sasin JM, Bujnicki JM (2003) A “Frankenstein’s monster” approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins 53 Suppl 6: 369–79.