Guettou F, Quistgaard EM, Raba M, Moberg P, Löw C, Nordlund P (2014) Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2. Nature Structural and Molecular Biology 21:728-731. doi: https://doi.org/10.1038/nsmb.2860
Dovega R, Tsutakawa S, Quistgaard EM, Anandapadamanaban M, Löw C, Nordlund P (2014) Structural and biochemical characterization of human PR70 in isolation and in complex with the scaffolding subunit of protein phosphatase 2A. PLoS ONE 9:e101846. doi: https://doi.org/10.1371/journal.pone.0101846
Löw C, Quistgaard EM, Kovermann M, Anandapadamanaban M, Balbach J, Nordlund P (2014) Structural basis for PTPA interaction with the invariant C-terminal tail of PP2A. Biological Chemistry 395:881-889. doi: https://doi.org/10.1515/hsz-2014-0106,
2013
Löw C, Yau YH, Pardon E, Jegerschöld C, Wåhlin L, Quistgaard EM, Moberg P, Geifman-Shochat S, Steyaert J, Nordlund P (2013) Nanobody mediated crystallization of an archeal mechanosensitive channel. PLoS One, e77984.
Quistgaard EM, Löw C, Moberg P, Nordlund P Metal-mediated crystallization of the xylose transporter XylE from Escherichia coli in three different crystal forms. J Struct Biol. 184; 375-8.
Quistgaard EM, Löw C, Moberg P, Guettou F, Maddi K, Nordlund P (2013) Structural and biophysical characterization of the cytoplasmic domains of human BAP29 and BAP31. (2013) PLoS One, e71111.
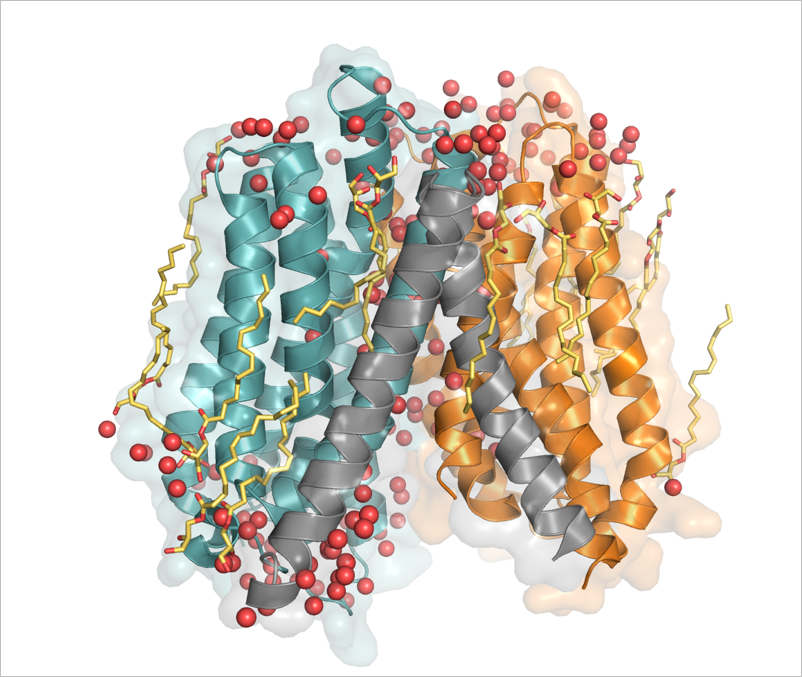
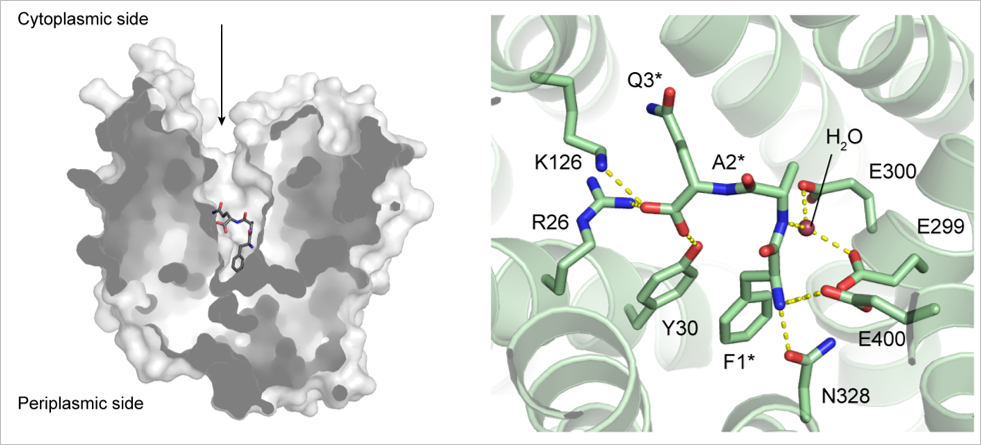
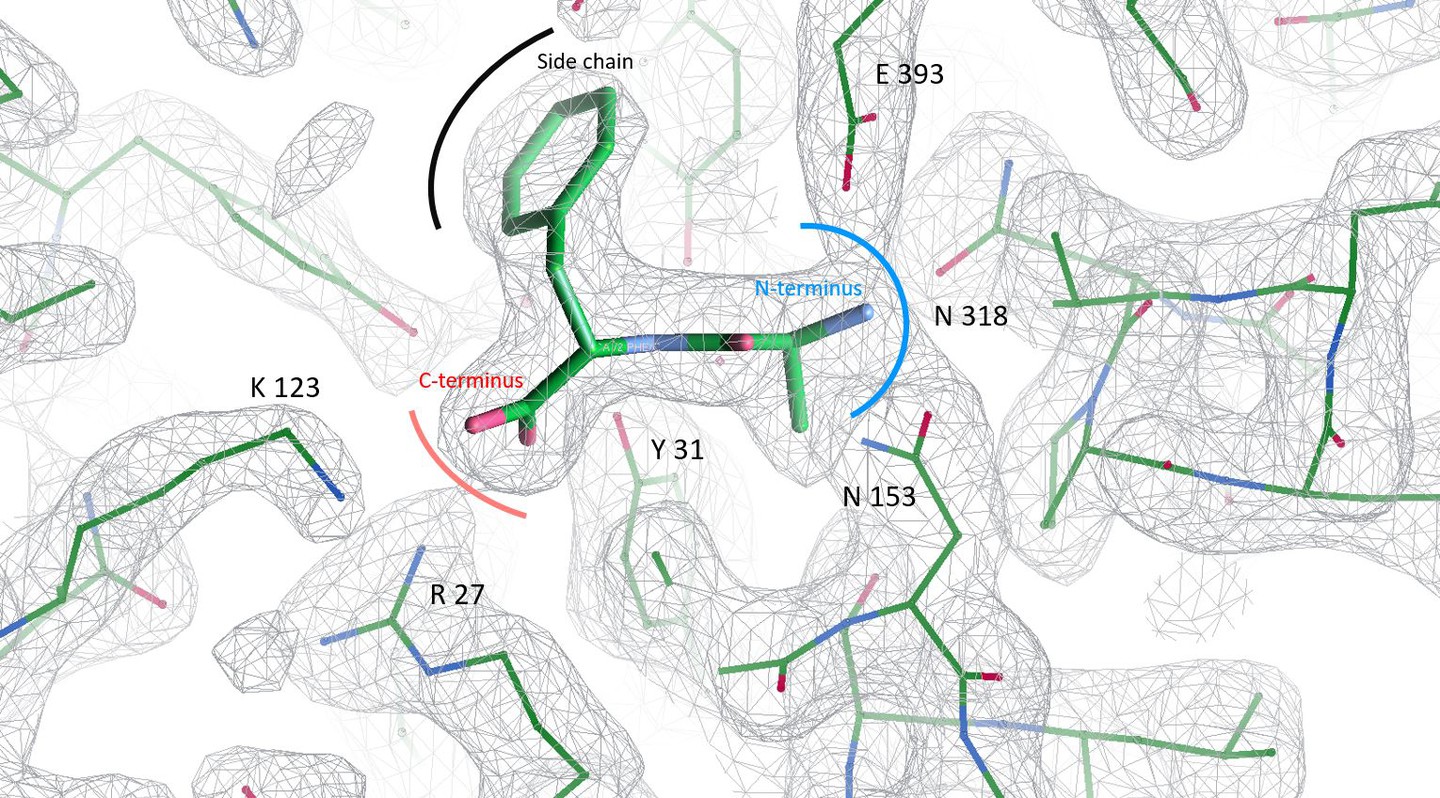
Guettou F, Quistgaard EM, Trésaugues L, Moberg P, Jegerschöld C, Zhu L, Jong AJ, Nordlund P, Löw C (2013) Structural insights into substrate recognition in proton-dependent oligopeptide transporters. EMBO Rep 14, 804-10.
Weininger U, Respondek M, Löw C, Akke M (2013) Slow aromatic ring flips detected despite near-degenerate NMR frequencies of the exchanging nuclei. J Phys Chem B 117, 9241-7.
Quistgaard EM, Löw C, Moberg P, Tresaugues L, Nordlund P (2013) Structural basis for substrate translocation in the GLUT homology family of monosaccharide transporters. Nature Structural and Molecular Biology 20, 766-8.
Löw C, Moberg P, Quistgaard EM, Hedrén M, Guettou F, Frauenfeld Haneskog L, Nordlund P (2013) High-throughput analytical gel filtration screening of integral membrane proteins for structural studies. Biochim Biophys Acta. 1830, 3497-508.
Guettou F, Löw C, Quistgaard EM, Hedrén M, Frauenfeld J, Nordlund P, Haneskog L, Moberg P. (2013) Rapid screening of integral membrane proteins suitable for structure determination. Discovery Matters 16
2012
Quistgaard EM, Nordlund P, Löw C (2012) High-resolution insights into binding of unfolded polypeptides by the PPIase chaperone SlpA. FASEB Journal 26, 4003-4013.
Löw C, Jegerschöld C, Kovermann M, Moberg P, Nordlund P # (2012) Optimisation of over-expression and biophysical characterisation of human membrane protein synaptogyrin 1. PLoS One, e38244.
Löw C, Stubbs MT, Haupt C, Balbach J (2012) Metallochaperone SlyD. Encyclopedia of Inorganic and Bioinorganic Chemistry. https://doi.org/10.1002/9781119951438.eibc2061
2011
Klepsch M, Kovermann M, Löw C, Balbach J, Permentier HP, Fusetti F, de Gier JW, Slotboom DJ, Berntsson RP (2011)Escherichia coli binding protein OppA has a preference for positively charged peptides. Journal of Molecular Biology 414, 75-85
Kahra D, Kovermann M, Löw C, Hirschfeld V, Haupt C, Balbach J, Hübner C (2011) Conformational plasticity and dynamics in a generic protein folding catalyst unraveled by single-molecule FRET. Journal of Molecular Biology 411, 781-90
Kovermann M, Zierold R, Haupt C, Löw C, Balbach J (2011) NMR relaxation unravels interdomain crosstalk of the two domain prolyl isomerase and chaperone SlyD. Biochim. Biophys. Acta. 1814, 873-81
Moberg P, Burstedt M, Nordlund P, Haneskog L, Löw C (2011) Stability of membrane proteins analyzed by gel filtration. Discovery Matters (13)
Löw C, Hedrén M, Nordlund P, Haneskog L, Moberg P (2011) Rapid buffer scouting and quality control of integral membrane proteins. Discovery Matters (12):10-11.
2010
Löw C, Neumann P, Tidow H, Weininger U, Haupt C, Friedrich-Epler B, Scholz C, Stubbs MT, Balbach J (2010) Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus. Journal of Molecular Biology 398, 375-390
2009
Weininger U, Zeeb M, Neumann P, Löw C, Stubbs MT, Lipps G, Balbach J (2009) Structure based stability analysis of an extremely stable dimeric DNA binding protein from Sulfolobus islandicus. Biochemistry 48, 10030-7
Schulenburg C, Löw C, Weininger U, Mrestani-Klaus C, Hofmann H, Balbach J, Ulbrich-Hofmann R, Arnold U (2009) The folding pathway of Onconase is directed by a conserved intermediate. Biochemistry 48, 8449-57
Hofmann H, Weininger U, Löw C, Golbik RP, Balbach J, Ulbrich-Hofmann R (2009) Fast amide proton exchange reveals close relation between native-state dynamics and unfolding kinetics. J Am Chem Soc. 14, 140-146
Löw C, Homeyer N, Weininger U, Sticht H, Balbach J (2009) Conformational Switch upon Phosphorylation: Human CDK Inhibitor p19INK4d Between Native and Intermediate State. ACS Chem Biol. 16, 53-63 (2009); PMID: 19063602. – (“Pionts of view”- Doug Barrick. Biological Regulation via Ankyrin Repeat Folding. ACS Chem Biol. 16, 19-22
Löw C (2009) Energy Landscapes of Protein Folding: From Structure to Function. SVH Verlag, Saarbrücken.
2008
Löw C, Weininger U, Lee H, Schweimer K, Neundorf I, Beck-Sickinger AG, Pastor RW, Balbach J (2008) Structure and Dynamics of Helix-0 of the N-BAR Domain in Lipid Micelles and Bilayers. Biophys J. 95, 4315-23
Löw C, Weininger C, Neumann P, Klepsch M, Lilie H, Stubbs MT, Balbach J (2008) Structural Insights into an Equilibrium Folding Intermediate of an Archaeal Ankyrin Repeat Protein. Proc. Natl. Acad. Sci. 105, 3779-3784
2007
Schulenburg C, Martinez-Senac MM, Löw C, Golbik RP, Ulbrich-Hofmann R, Arnold U (2007) Identification of three Phases in Onconase Refolding. FEBS Journal 274, 5826-5833
Löw C, Weininger U, Zeeb M, Zhang W, Laue ED, Schmid FX, Balbach J (2007) Folding Mechanism of an Ankyrin Repeat Protein: Scaffold and Active Site Formation of Human CDK Inhibitor p19INK4d. Journal of Molecular Biology 373, 219-231 (Faculty of 1000 - evaluation)
2006
Zeeb M, Max KEA, Weininger U, Löw C, Sticht H, Balbach, J (2006)Recognition of T-rich single-stranded DNA by the Cold Shock Protein Bs-CspB in Solution. Nucl. Acids Res. 34, 4561-4571