Research Projects
PREVIOUS AND CURRENT RESEARCH
The order of Bunyavirales is a highly diverse group of RNA viruses including numerous human pathogens. The WHO R&D Blueprint emphasizes the urgent need for medical countermeasures against several bunyaviruses, including the Lassa virus, Crimean-Congo hemorrhagic fever virus and Rift Valley fever virus. These viruses are a threat to global health and the economy especially in low- and middle-income countries but they are also recognized for their high epidemic potential and, due to climate change, their expanding geographic distribution. Many of these viruses must be handled in biocontainments of the highest biosafety standards limiting the method and technology spectrum for research. Despite the severe diseases they can cause, bunyaviruses are yet fascinatingly simple: they contain a segmented negative-strand RNA genome and produce only a handful of own proteins, some of them get by with only four gene products.
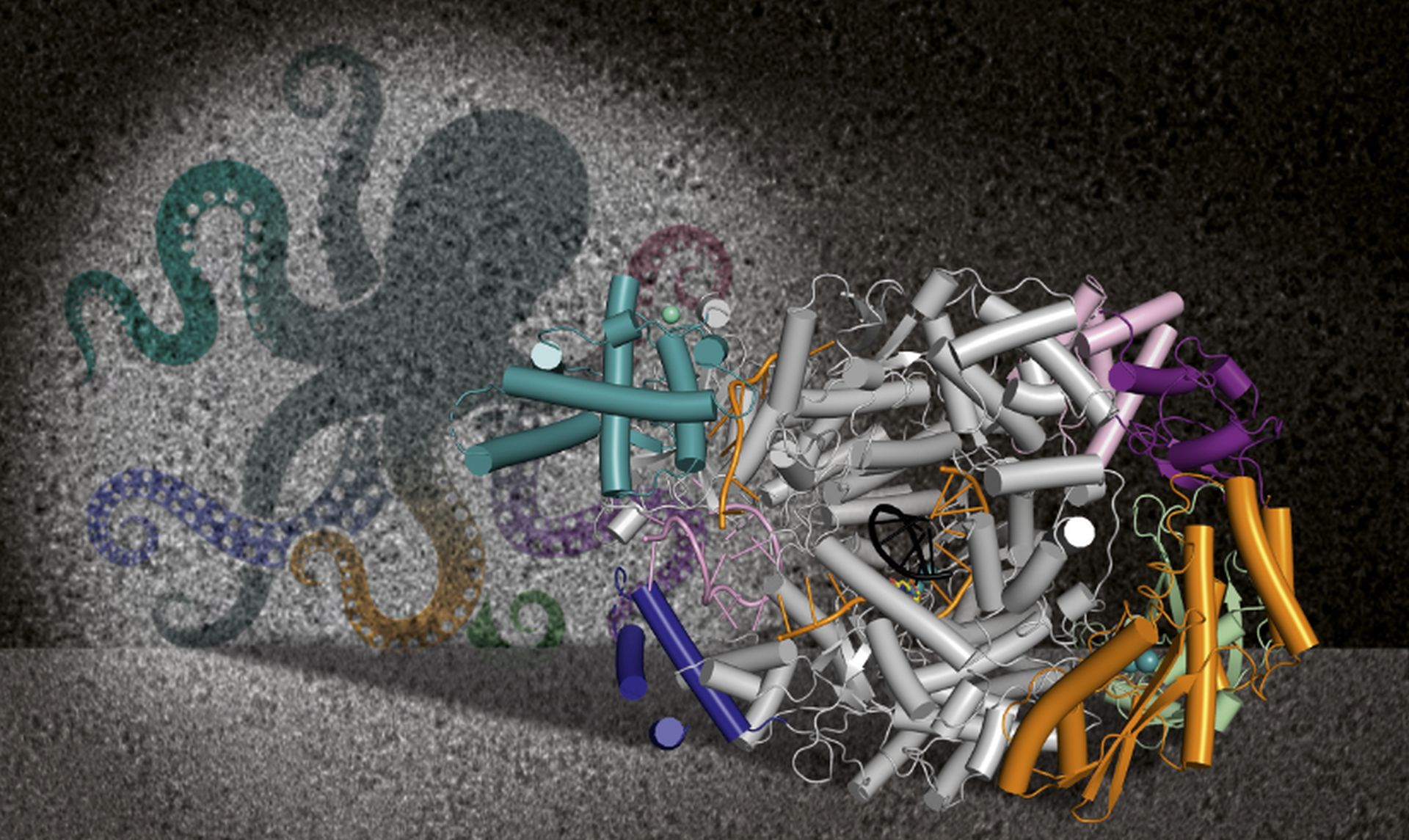
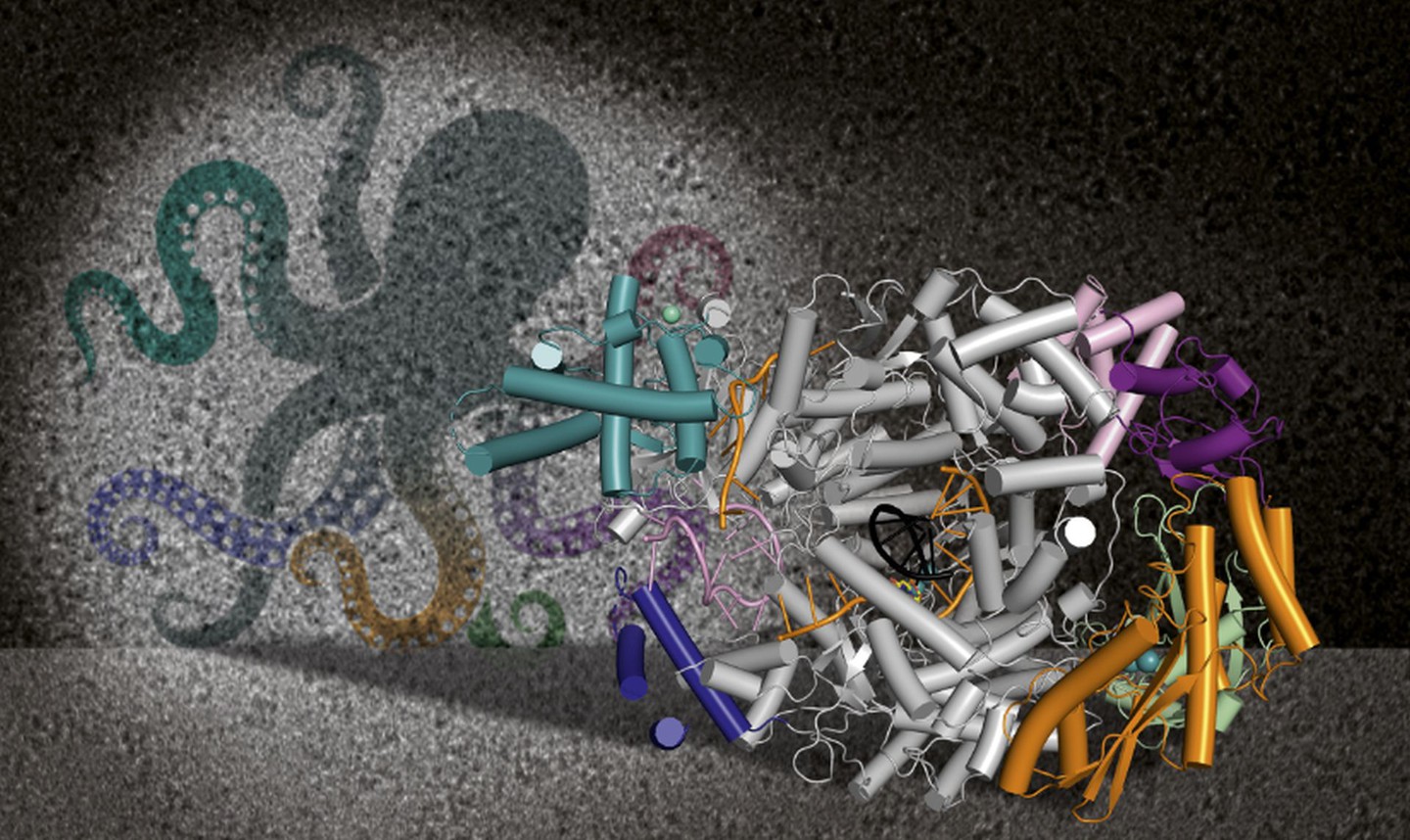
Two proteins are mainly in the focus of our research: The large (L) protein and the nucleoprotein N that together with the RNA genome of the virus form the ribonucleoparticle, which is sufficient for replication and transcription of the viral genome. We apply a variety of methods from structural biology, biochemistry and virology fields to understand bunyavirus genome replication and transcription processes in tiny detail. These processes lead to production of new viral proteins in cells (viral transcription) as well as the amplification of the viral genome (genome replication) to finally assemble new viral particles, ready to infect the next cells. The key player in these processes is the viral L protein, which is a multifunctional and highly optimized molecular machine, comparable to a swiss army knife. Our goal is to understand this molecular machine - its functions, its regulation, its interactors - to finally develop strategies to inhibit the processes of genome replication and transcription and thereby stop bunyavirus amplification.
We focus on several key aspects of bunyavirus genome replication and transcription:
- Structural and functional characterization of the L protein. In the past years we have determined several L protein structures of different bunyaviruses, specifically Lassa virus (family Arenaviridae), severe fever with thrombocytopenia syndrome virus (family Phenuiviridae) and Sin Nombre virus (family Hantaviridae), via single-particle cryoEM in close collaboration with the Grünewald lab as well as Stephen Cusack (EMBL Grenoble). We developed in vitro assays to characterize the different functions of L and define the optimal conditions to stall the protein in functional states of genome replication and transcription for cryoEM imaging. We visualized the interaction of L with the RNA termini of the viral genome, the active site of L during active RNA synthesis, autoinhibition of enzymatic activities as well as an enormous conformational flexibility of this multidomain protein.
- We use X-ray crystallography to solve high-resolution structures of isolated L protein domains, such as the cap-binding domains and the endonuclease. These data can be intergrated with cryoEM and SAXS data of the full-length L protein as these domains are highly flexible and often rather poorly defined in the cryoEM maps. As both domains are needed for the virus-specific cap-snatching mechanism to prime viral transcription, their structures are directly relevant for drug development purposes.
- The structural and functional insights into L protein function and regulation are used to design robust, high-throughput-feasible assay systems and screen large compound libraries in collaboration with the Fraunhofer Institute for Translational Medicine and Pharmacology, ScreeningPort Hamburg.
- Using native mass spectrometry (nMS) and hydrogen-deuterium exchange mass spectrometry (HDX-MS), together with the Uetrecht lab we investigate the interactions between viral proteins, viral proteins and viral RNA as well as between viral and host cell proteins. The molecular characterization of direct interactions again offers possibilities for new antiviral strategies.
- An additional research topic in the past years has been virus-host interactions. We employed screening approaches combined with a bioinformatic systems approach in collaboration with the Kosinski lab. We are currently following up several interesting candidates by cell-based and in vitro experiments.
FUTURE PROJECTS AND GOALS
- A detailed molecular understanding of the complete processes of bunyavirus genome replication and transcription
- Structural and functional characterization of host protein-L protein complexes
- Identification of further druggable targets for antiviral drug development
SELECTED PUBLICATIONS
Meier K, Thorkelsson SR, Trouilleton GD, Vogel D, Yu D, Kosinski J, Cusack S, Malet H, Grünewald K, Quemin ERJ, Rosenthal M. Structural and functional characterization of the Sin Nombre virus L protein. Submitted February 2023
Saenger L, Williams HM, Yu D., Vogel D. Kosinski J, Rosenthal M*, Uetrecht C* (2023) An RNA to rule them all: Critical steps in Lassa virus ribonucleoparticle assembly and recruitment. BioRxiv DOI: 10.1101/2023.02.09.527830
Williams HM, Thorkelsson SR, Vogel D, Milewski M, Busch C, Cusack S, Grünewald K, Quemin ERJ, Rosenthal M (2023) Structural insights into viral genome replication by the Severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Res. 51(3):1424-1442. doi: 10.1093/nar/gkac1249.
Yu D, Chojnowski G, Rosenthal M, Kosinski J (2023) AlphaPulldown – a Python package for protein-protein interaction screens using AlphaFold-Multimer. Bioinformatics39(1):btac749. doi: 10.1093/bioinformatics/btac749.
Soh TK, Pfefferle S, Wurr S, von Possel R, Oestereich L, Rieger T, Rosenthal M*, Bosse JB* (2023) A validated protocol to UV-inactivate SARS-CoV-2 and herpesvirus-infected cells. PLoS One. 18(5):e0274065. doi: 10.1371/journal.pone.0274065.
Kouba T, Vogel D, Thorkelsson SR, Quemin E, Williams HM, Milewski M, Busch C, Günther S, Grünewald K, Rosenthal M*, Cusack S*(2021) Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity. Nat Commun. 12(1):7018. doi: 10.1038/s41467-021-27305-5.
Cusack S, Rosenthal M (2021) Matters arising: Errors in the deposited SFTSV L protein structure (PDB:6L42). Nat Microbiol. 6(5):549-550. doi: 10.1038/s41564-021-00901-3.
Vogel D, Thorkelsson SR, Quemin E, Meier K, Kouba T, Gogrefe N, Busch C, Reindl S, Günther S, Cusack S, Grünewald K, Rosenthal M (2020) Structural and functional characterization of the Severe fever with thrombocytopenia syndrome virus L protein. Nucleic Acids Res. 48(10):5749-5765. doi: 10.1093/nar/gkaa253.
Jones R, Lessoued S, Meier K, Devignot S, Barata S, Mate M, Bragagnolo G, Weber F, Rosenthal M, Reguera J (2019) Structure and function of the Toscana virus cap-snatching endonuclease. Nucleic Acids Res. 47(20):10914-10930. doi: 10.1093/nar/gkz838.
Gogrefe N, Reindl S, Günther S, Rosenthal M (2019) Structure of a functional cap-binding domain in Rift Valley fever virus L protein. PLoS Pathog. 15(5):e1007829. doi: 10.1371/journal.ppat.1007829.
Jérôme H, Rudolf M, Lelke M, Pahlmann M, Busch C, Bockholt S, Wurr S, Günther S, Rosenthal M*, Kerber R* (2019) Rift Valley fever virus minigenome system for investigating the role of L protein residues in viral transcription and replication. J Gen Virol. 100(7):1093-1098. doi: 10.1099/jgv.0.001281.
Vogel D*, Rosenthal M*, Gogrefe N, Reindl S, Günther S (2019) Biochemical characterization of the Lassa virus L protein. J Biol Chem. 294(20):8088-8100. doi: 10.1074/jbc.RA118.006973.