Publications
Preprints
Genz LR, Nair S, Topf M (2025) Assessing scoring metrics for AlphaFold2 and AlphaFold3 protein complex predictions. bioRxiv 2025.04.16.648930; doi: 10.1101/2025.04.16.648930
Topf M, Maiorca M, Jadhav S, Sweeney A, Marini G, Mulvaney T, Marcia M (2025) Uncovering Hidden Functional States in Cryo-EM Datasets with EMPROVE, Research Square; https://doi.org/10.21203/rs.3.rs-6535769/v1
Marcia M, Jadhav S, Maiorca M, Manigrasso J, Muscat S, Mulvaney T, De Vivo M, Topf M (2025) Dynamic assembly of a large multidomain ribozyme visualized by cryo-electron microscopy, Research Square; https://doi.org/10.21203/rs.3.rs-5664507/v1
Cook AD, Roberts AJ, Atherton J, Tewari R, Topf M, Moores CA (2021) Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition. J Biol Chem. 101063. doi: 10.1016/j.jbc.2021.101063.
Malhotra S, Joseph A-P, Topf M (2021) Using known components or homologs: model building. In: Glaeser RM, Nogales E, Chiu W (eds) Single-particle Cryo-EM of Biological Macromolecules, Biophysica. IOP Publishing, Bristol, UK, pp 244–247
Peña A, Sweeney A, Cook AD, Locke J, Topf M, Moores CA (2021) Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy. Structure. 28(4):450-457.e5. doi: 10.1016/j.str.2020.01.013
Cragnolini T, Sweeney A, Topf M (2021b) Automated Modeling and Validation of Protein Complexes in Cryo-EM Maps. Methods Mol Biol 2215:189–223. https://doi.org/10.1007/978-1-0716-0966-8_9
2020
Cif L, Demailly D, Lin J-P, et al (2020) KMT2B-related disorders: expansion of the phenotypic spectrum and long-term efficacy of deep brain stimulation. Brain. https://doi.org/10.1093/brain/awaa304
Joseph AP, Lagerstedt I, Jakobi A, Burnley T, Patwardhan A, Topf M, Winn M (2020) Comparing Cryo-EM Reconstructions and Validating Atomic Model Fit Using Difference Maps. J Chem Inf Model 60:2552–2560. https://doi.org/10.1021/acs.jcim.9b01103
Peña A, Sweeney A, Cook AD, Locke J,Topf M, Moores CA (2020) Structure of Microtubule-Trapped Human Kinesin-5 and Its Mechanism of Inhibition Revealed Using Cryoelectron Microscopy. Structure 28:450-457.e5. https://doi.org/10.1016/j.str.2020.01.013
Sinnott M, Malhotra S, Madhusudhan MS, Thalassinos K, Topf M (2020) Combining Information from Crosslinks and Monolinks in the Modeling of Protein Structures. Structure 28:1061-1070.e3. https://doi.org/10.1016/j.str.2020.05.012
Vollmer B, Pražák V, Vasishtan D, Jefferys EE , Hernandez-Duran A, Vallbracht M, Klupp B, Mettenleiter TC, Backovic M, Rey FA, Topf M, Grünewald K (2020) The prefusion structure of herpes simplex virus glycoprotein B. Sci Adv Adv 6:1–11
2019
Carecchio M, Invernizzi F, Gonzàlez-Latapi P, et al (2019) Frequency and phenotypic spectrum of KMT2B dystonia in childhood: A single-center cohort study. Mov Disord 34:1516–1527. https://doi.org/10.1002/mds.27771
Demetriou C, Chanudet E, Joseph A, et al (2019) Exome sequencing identifies variants in FKBP4 that are associated with recurrent fetal loss in humans. Hum Mol Genet 28:3466–3474. https://doi.org/10.1093/hmg/ddz203
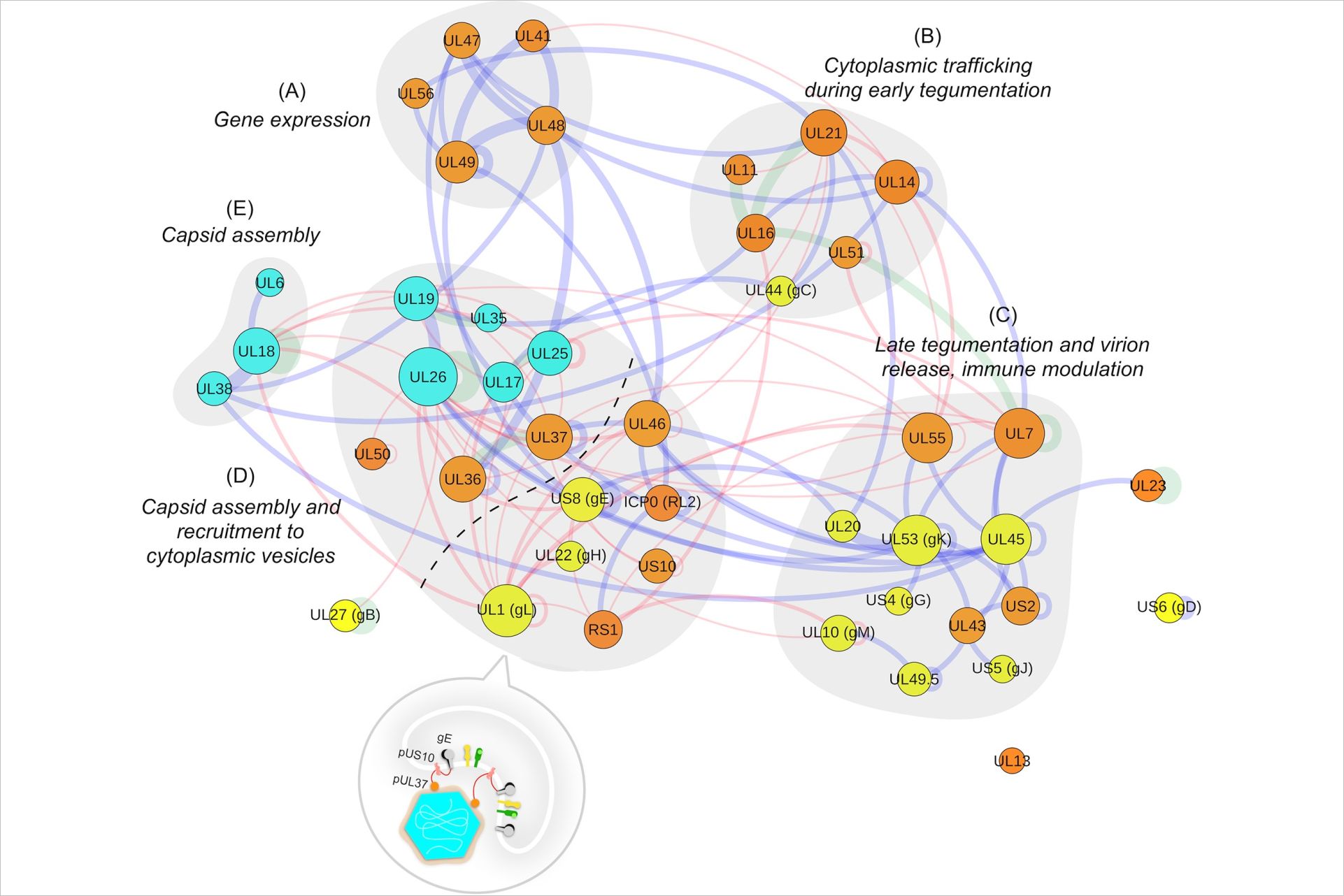
Hernández Durán A, Greco TM, Vollmer B, et al (2019a) Protein interactions and consensus clustering analysis uncover insights into herpesvirus virion structure and function relationships. PLoS Biol 17:. https://doi.org/10.1371/journal.pbio.3000316
Hernández Durán A, Grünewald K, Topf M (2019b) Conserved Central Intraviral Protein Interactome of the Herpesviridae Family. mSystems 4:. https://doi.org/10.1128/msystems.00295-19
Ignatiou A, Brasilès S, El Sadek Fadel M, et al (2019) Structural transitions during the scaffolding-driven assembly of a viral capsid. Nat Commun 10:4840. https://doi.org/10.1038/s41467-019-12790-6
Kryshtafovych A, Malhotra S, Monastyrskyy B, et al (2019a) Cryo-electron microscopy targets in CASP13: Overview and evaluation of results. Proteins Struct Funct Bioinforma 87:1128–1140. https://doi.org/10.1002/prot.25817
Kryshtafovych A, Schwede T, Topf M, et al (2019b) Critical assessment of methods of protein structure prediction (CASP)—Round XIII. Proteins Struct. Funct. Bioinforma. 87:1011–1020
Lepore R, Kryshtafovych A, Alahuhta M, et al (2019) Target highlights in CASP13: Experimental target structures through the eyes of their authors. Proteins Struct Funct Bioinforma 87:1037–1057. https://doi.org/10.1002/prot.25805
Malhotra S, Träger S, Dal Peraro M, Topf M (2019) Modelling structures in cryo-EM maps. Curr. Opin. Struct. Biol. 58:105–114
Pereira R, Gendron T, Sanghera C, et al (2019) Mapping Aldehyde Dehydrogenase 1A1 Activity using an [18 F]Substrate-Based Approach. Chem - A Eur J 25:2345–2351. https://doi.org/10.1002/chem.201805473
2018
Al-Olabi L, Polubothu S, Dowsett K, et al (2018) Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J Clin Invest 128:1496–1508. https://doi.org/10.1172/JCI98589
Bullock JMA, Sen N, Thalassinos K, Topf M (2018a) Modeling Protein Complexes Using Restraints from Crosslinking Mass Spectrometry. Structure 26:1015-1024.e2. https://doi.org/10.1016/j.str.2018.04.016
Bullock JMA, Thalassinos K, Topf M (2018b) Jwalk and MNXL web server: Model validation using restraints from crosslinking mass spectrometry. Bioinformatics 34:3584–3585. https://doi.org/10.1093/bioinformatics/bty366
Fedele L, Newcombe J, Topf M, et al (2018) Disease-associated missense mutations in GluN2B subunit alter NMDA receptor ligand binding and ion channel properties. Nat Commun 9:. https://doi.org/10.1038/s41467-018-02927-4
Fumagalli M, Camus SM, Diekmann Y, et al (2018) Genetic diversity of CHC22 clathrin impacts its function in glucose metabolism. bioRxiv 307264
McTague A, Nair U, Malhotra S, et al (2018) Clinical and molecular characterization of KCNT1-related severe early-onset epilepsy. Neurology 90:E55–E66. https://doi.org/10.1212/WNL.0000000000004762
Menny A, Serna M, Boyd CM, et al (2018) CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers. bioRxiv
Newcombe J, Chatzidaki A, Sheppard TD, et al (2018) Diversity of nicotinic acetylcholine receptor positive allosteric modulators revealed by mutagenesis and a revised structural model. Mol Pharmacol 93:128–140. https://doi.org/10.1124/mol.117.110551
Tweedy JG, Escriva E, Topf M, Gompels UA (2018) Analyses of tissue culture adaptation of human herpesvirus-6a by whole genome deep sequencing redefines the reference sequence and identifies virus entry complex changes. Viruses. https://doi.org/10.3390/v10010016
Smelt CLC, Sanders VR, Newcombe J, et al (2018) Identification by virtual screening and functional characterisation of novel positive and negative allosteric modulators of the α7 nicotinic acetylcholine receptor. Neuropharmacology 139:194–204. https://doi.org/10.1016/j.neuropharm.2018.07.009
Pilorge M, Fassier C, Le Corronc H, et al (2016) Genetic and functional analyses demonstrate a role for abnormal glycinergic signaling in autism. Mol Psychiatry 21:936–945. https://doi.org/10.1038/mp.2015.139
2017
Atherton J, Jiang K, Stangier MM, et al (2017a) A structural model for microtubule minus-end recognition and protection by CAMSAP proteins. Nat Struct Mol Biol 24:931–943. https://doi.org/10.1038/nsmb.3483
Atherton J, Yu IM, Cook A, et al (2017b) The divergent mitotic kinesin MKLP2 exhibits atypical structure and mechanochemistry. Elife 6:. https://doi.org/10.7554/eLife.27793
Deville C, Carroni M, Franke KB, et al (2017) Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci Adv 3:e1701726. https://doi.org/10.1126/sciadv.1701726
Joseph AP, Lagerstedt I, Patwardhan A, et al (2017a) Improved metrics for comparing structures of macromolecular assemblies determined by 3D electron-microscopy. J Struct Biol 199:12–26. https://doi.org/10.1016/j.jsb.2017.05.007
Joseph AP, Polles G, Alber F, Topf M (2017b) Integrative modelling of cellular assemblies. Curr. Opin. Struct. Biol. 46:102–109
Locke J, Joseph AP, Peña A, et al (2017) Structural basis of human kinesin-8 function and inhibition. Proc Natl Acad Sci U S A 114:E9539–E9548. https://doi.org/10.1073/pnas.1712169114
Meyer E, Carss KJ, Rankin J, et al (2017) Mutations in the histone methyltransferase gene KMT2B cause complex early-onset dystonia. Nat Genet 49:223–237. https://doi.org/10.1038/ng.3740
Schaefer N, Berger A, Van Brederode J, et al (2017) Disruption of a structurally important extracellular element in the glycine receptor leads to decreased synaptic integration and signaling resulting in severe startle disease. J Neurosci 37:7948–7961. https://doi.org/10.1523/JNEUROSCI.0009-17.2017
Wright ZVF, McCarthy S, Dickman R, et al (2017) The Role of Disulfide Bond Replacements in Analogues of the Tarantula Toxin ProTx-II and Their Effects on Inhibition of the Voltage-Gated Sodium Ion Channel Nav1.7. J Am Chem Soc 139:13063–13075. https://doi.org/10.1021/jacs.7b06506
2016
Ashford P, Hernandez A, Greco TM, et al (2016) HVint: A strategy for identifying novel protein-protein interactions in herpes simplex virus type 1. Mol Cell Proteomics 15:2939–2953. https://doi.org/10.1074/mcp.M116.058552
Bullock JMA, Schwab J, Thalassinos K, Topf M (2016) The importance of non-accessible crosslinks and solvent accessible surface distance in modeling proteins with restraints from crosslinking mass spectrometry. Mol Cell Proteomics 15:2491–2500. https://doi.org/10.1074/mcp.M116.058560
Joseph AP, Malhotra S, Burnley T, et al (2016) Refinement of atomic models in high resolution EM reconstructions using Flex-EM and local assessment. Methods 100:42–49. https://doi.org/10.1016/j.ymeth.2016.03.007
Kalscheuer VM, James VM, Himeiright ML, et al (2016) Novel missense mutation A789V in IQSEC2 underlies X-linked intellectual disability in the MRX78 family. Front Mol Neurosci 8:85. https://doi.org/10.3389/fnmol.2015.00085
Zeev-Ben-Mordehai T, Vasishtan D, Durán AH, et al (2016) Two distinct trimeric conformations of natively membrane-anchored full-length herpes simplex virus 1 glycoprotein B. Proc Natl Acad Sci U S A 113:4176–4181. https://doi.org/10.1073/pnas.1523234113
2015
Farabella I, Vasishtan D, Joseph AP, et al (2015) TEMPy: A Python library for assessment of three-dimensional electron microscopy density fits. J Appl Crystallogr 48:1314–1323. https://doi.org/10.1107/S1600576715010092
Wood C, Burnley T, Patwardhan A, et al (2015) Collaborative computational project for electron cryo-microscopy. Acta Crystallogr Sect D Biol Crystallogr 71:123–126. https://doi.org/10.1107/S1399004714018070
Stödberg T, McTague A, Ruiz AJ, et al (2015) Mutations in SLC12A5 in epilepsy of infancy with migrating focal seizures. Nat Commun 6:. https://doi.org/10.1038/ncomms9038
Pandurangan AP, Vasishtan D, Alber F, Topf M (2015) γ-TEMPy: Simultaneous Fitting of Components in 3D-EM Maps of Their Assembly Using a Genetic Algorithm. Structure 23:2365–2376. https://doi.org/10.1016/j.str.2015.10.013
Lukoyanova N, Kondos SC, Farabella I, et al (2015) Conformational Changes during Pore Formation by the Perforin-Related Protein Pleurotolysin. PLoS Biol 13:. https://doi.org/10.1371/journal.pbio.1002049
Sali A, Berman HM, Schwede T, et al (2015) Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. In: Structure. Cell Press, pp 1156–1167
2014
Atherton J, Farabella I, Yu IM, et al (2014) Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. Elife 3:e03680. https://doi.org/10.7554/eLife.03680
Leung C, Dudkina N V., Lukoyanova N, et al (2014) Stepwise visualization of membrane pore formation by suilysin, a bacterial cholesterol-dependent cytolysin. Elife 3:e04247. https://doi.org/10.7554/eLife.04247
Mortensen M, Iqbal F, Pandurangan AP, et al (2014) Photo-antagonism of the GABA A receptor. Nat Commun 5:. https://doi.org/10.1038/ncomms5454
Pandurangan AP, Shakeel S, Butcher SJ, Topf M (2014) Combined approaches to flexible fitting and assessment in virus capsids undergoing conformational change. J Struct Biol 185:427–439. https://doi.org/10.1016/j.jsb.2013.12.003
2013
Ganser LR, Yan Q, James VM, et al (2013) Distinct phenotypes in zebrafish models of human startle disease. Neurobiol Dis 60:139–151. https://doi.org/10.1016/j.nbd.2013.09.002
James VM, Bode A, Chung SK, et al (2013) Novel missense mutations in the glycine receptor β subunit gene (GLRB) in startle disease. Neurobiol Dis 52:137–149. https://doi.org/10.1016/j.nbd.2012.12.001
Maurer UE, Zeev-Ben-Mordehai T, Pandurangan AP, et al (2013) The structure of herpesvirus fusion glycoprotein B-bilayer complex reveals the protein-membrane and lateral protein-protein interaction. Structure 21:1396–1405. https://doi.org/10.1016/j.str.2013.05.018
Yuan S, Topf M, Reubold TF, et al (2013) Changes in apaf-1 conformation that drive apoptosome assembly. Biochemistry 52:2319–2327. https://doi.org/10.1021/bi301721g
Redzej A, Ilangovan A, Lang S, et al (2013) Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems. Mol Microbiol 89:324–333. https://doi.org/10.1111/mmi.12275
Thalassinos K, Pandurangan AP, Xu M, et al (2013) Conformational states of macromolecular assemblies explored by integrative structure calculation. Structure 21:1500–1508
2012
Carta E, Chung SK, James VM, et al (2012) Mutations in the GlyT2 gene (SLC6A5) are a second major cause of startle disease. J Biol Chem 287:28975–28985. https://doi.org/10.1074/jbc.M112.372094
Clare DK, Vasishtan D, Stagg S, et al (2012) ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell 149:113–123. https://doi.org/10.1016/j.cell.2012.02.047
Gill JL, James VM, Carta E, et al (2012) Identification of congenital muscular dystonia 2 associated with an inherited GlyT2 defect in Belgian Blue cattle from the United Kingdom. Anim Genet 43:267–270. https://doi.org/10.1111/j.1365-2052.2011.02255.x
Giménez C, Pérez-Siles G, Martínez-Villarreal J, et al (2012) A novel dominant hyperekplexia mutation Y705C alters trafficking and biochemical properties of the presynaptic glycine transporter GlyT2. J Biol Chem 287:28986–29002. https://doi.org/10.1074/jbc.M111.319244
James VM, Gill JL, Topf M, Harvey RJ (2012) Molecular mechanisms of glycine transporter GlyT2 mutations in startle disease. In: Biological Chemistry. pp 283–289
Pandurangan AP, Topf M (2012a) RIBFIND: A web server for identifying rigid bodies in protein structures and to aid flexible fitting into cryo EM maps. Bioinformatics 28:2391–2393. https://doi.org/10.1093/bioinformatics/bts446
Pandurangan AP, Topf M (2012b) Finding rigid bodies in protein structures: Application to flexible fitting into cryoEM maps. J Struct Biol 177:520–531. https://doi.org/10.1016/j.jsb.2011.10.011
Seitsonen JJT, Shakeel S, Susi P, et al (2012) Structural Analysis of Coxsackievirus A7 Reveals Conformational Changes Associated with Uncoating. J Virol 86:7207–7215. https://doi.org/10.1128/jvi.06425-11
2011
Beck M, Topf M, Frazier Z, et al (2011) Exploring the spatial and temporal organization of a cell’s proteome. J Struct Biol 173:483–496. https://doi.org/10.1016/j.jsb.2010.11.011
Vasishtan D, Topf M (2011) Scoring functions for cryoEM density fitting. J Struct Biol 174:333–343. https://doi.org/10.1016/j.jsb.2011.01.012
Yuan S, Yu X, Topf M, et al (2011) Structure of the drosophila apoptosome at 6.9 Å resolution. Structure 19:128–140. https://doi.org/10.1016/j.str.2010.10.009
2010
Gartmann M, Blau M, Armache JP, et al (2010) Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem 285:14848–14851. https://doi.org/10.1074/jbc.C109.096057
Kumar RA, Pilz DT, Babatz TD, et al (2010) TUBA1A mutations cause wide spectrum lissencephaly (smooth brain) and suggest that multiple neuronal migration pathways converge on alpha tubulins. Hum Mol Genet 19:2817–2827. https://doi.org/10.1093/hmg/ddq182
Yuan S, Yu X, Topf M, et al (2010) Structure of an apoptosome-procaspase-9 CARD complex. Structure 18:571–583. https://doi.org/10.1016/j.str.2010.04.001
Zhang S, Vasishtan D, Xu M, et al (2010) A fast mathematical programming procedure for simultaneous fitting of assembly components into cryoEM density maps. Bioinformatics 26:i261–i268. https://doi.org/10.1093/bioinformatics/btq201
Malet H, Topf M, Clare DK, et al (2010) RNA channelling by the eukaryotic exosome. EMBO Rep 11:936–942. https://doi.org/10.1038/embor.2010.164
Rawi R, Whitmore L, Topf M (2010) CHOYCE: A web server for constrained homology modelling with cryoEM maps. Bioinformatics 26:1673–1674. https://doi.org/10.1093/bioinformatics/btq237
2009
Lasker K, Topf M, Sali A, Wolfson HJ (2009) Inferential Optimization for Simultaneous Fitting of Multiple Components into a CryoEM Map of Their Assembly. J Mol Biol 388:180–194. https://doi.org/10.1016/j.jmb.2009.02.031
Taylor DJ, Devkota B, Huang AD, et al (2009) Comprehensive Molecular Structure of the Eukaryotic Ribosome. Structure 17:1591–1604. https://doi.org/10.1016/j.str.2009.09.015
2008
Alber F, Förster F, Korkin D, et al (2008) Integrating diverse data for structure determination of macromolecular assemblies. Annu. Rev. Biochem. 77:443–477
Booth CR, Meyer AS, Cong Y, et al (2008) Mechanism of lid closure in the eukaryotic chaperonin TRiC/CCT. Nat Struct Mol Biol 15:746–753. https://doi.org/10.1038/nsmb.1436
Chandramouli P, Topf M, Ménétret JF, et al (2008) Structure of the Mammalian 80S Ribosome at 8.7 Å Resolution. Structure 16:535–548. https://doi.org/10.1016/j.str.2008.01.007
Cong Y, Topf M, Sali A, et al (2008) Crystallographic Conformers of Actin in a Biologically Active Bundle of Filaments. J Mol Biol 375:331–336. https://doi.org/10.1016/j.jmb.2007.10.027
Connell SR, Topf M, Qin Y, et al (2008) A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol 15:910–915. https://doi.org/10.1038/nsmb.1469
Harvey RJ, Topf M, Harvey K, Rees MI (2008) The genetics of hyperekplexia: more than startle! Trends Genet. 24:439–447
Miller PS, Topf M, Smart TG (2008) Mapping a molecular link between allosteric inhibition and activation of the glycine receptor. Nat Struct Mol Biol 15:1084–1093. https://doi.org/10.1038/nsmb.1492
Serysheva II, Ludtke SJ, Baker ML, et al (2008) Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc Natl Acad Sci U S A 105:9610–9615. https://doi.org/10.1073/pnas.0803189105
Topf M, Lasker K, Webb B, et al (2008) Protein Structure Fitting and Refinement Guided by Cryo-EM Density. Structure 16:295–307. https://doi.org/10.1016/j.str.2007.11.016
2006
Topf M, Baker ML, Marti-Renom MA, et al (2006) Refinement of protein structures by iterative comparative modeling and cryoEM density fitting. J Mol Biol 357:1655–1668. https://doi.org/10.1016/j.jmb.2006.01.062
2005
Topf M, Baker ML, John B, et al (2005) Structural characterization of components of protein assemblies by comparative modeling and electron cryo-microscopy. J Struct Biol 149:191–203. https://doi.org/10.1016/j.jsb.2004.11.004
Topf M, Sali A (2005) Combining electron microscopy and comparative protein structure modeling. Curr. Opin. Struct. Biol. 15:578–585
2004
Topf M, Richards WG (2004) Theoretical studies on the deacylation step of serine protease catalysis in the gas phase, in solution, and in elastase. J Am Chem Soc 126:14631–14641. https://doi.org/10.1021/ja047010a
Topf M, Sandala GM, Smith DM, et al (2004) The unusual bifunctional catalysis of epimerization and desaturation by carbapenem synthase. J Am Chem Soc 126:9932–9933. https://doi.org/10.1021/ja047899v
Russell RB, Alber F, Aloy P, et al (2004) A structural perspective on protein-protein interactions. Curr. Opin. Struct. Biol. 14:313–324
Shacham S, Marantz Y, Bar-Haim S, et al (2004) PREDICT modeling and in-silico screening for G-protein coupled receptors. Proteins Struct Funct Genet 57:51–86. https://doi.org/10.1002/prot.20195
2003
Clifton IJ, Doan LX, Sleeman MC, et al (2003) Crystal structure of carbapenem synthase (CarC). J Biol Chem 278:20843–20850. https://doi.org/10.1074/jbc.M213054200
2002
Topf M, Várnai P, Richards WG (2002a) Ab initio QM/MM dynamics simulation of the tetrahedral intermediate of serine proteases: Insights into the active site hydrogen-bonding network. J Am Chem Soc 124:14780–14788. https://doi.org/10.1021/ja026219q
Topf M, Várnai P, Schofield CJ, Richards WG (2002b) Molecular dynamics simulations of the acyl-enzyme and the tetrahedral intermediate in the deacylation step of serine proteases. Proteins Struct Funct Genet 47:357–369. https://doi.org/10.1002/prot.10097
2001
Shacham S, Topf M, Avisar N, et al (2001) Modeling the 3D structure of GPCRs from sequence. Med Res Rev 21:472–483. https://doi.org/10.1002/med.1019
Topf M, Várnai P, Richards WG (2001) Quantum mechanical/molecular mechanical study of three stationary points along the deacylation step of the catalytic mechanism of elastase. Theor Chem Acc 106:146–151. https://doi.org/10.1007/s002140000246