Publications
Preprints
Maclot S, Reinert T, Duplantier L, Montagne G, Clavier C, Dagany X, Comby-Zerbino C, Kerleroux M, Thiede L, Pogan R, Uetrecht C, Kozhinov AN, Nagornov KO, Tsybin YO, Papanastasiou D, Antoine R (2025) Nanoelectrospray ionization coupled to a linear charge detection array ion trap spectrometer for single viral particles analysis. arXiv https://doi.org/10.48550/arXiv.2506.01593
Grün AFR, Said F-A, Schamoni-Kast K, Damjanovic T, Bosse J, Uetrecht C.(2025) Fast tracking native mass spectrometry: Skipping over buffer exchange. bioRxiv
https://doi.org/10.1101/2025.02.22.639503
Schamoni-Kast K, Uetrecht C. (2025) From Science to Fiction – Where are We Connecting in Vivo and in Vitro Results in Polyprotein Processing? SSRN http://dx.doi.org/10.2139/ssrn.5144731
Creon A, Scheer TES, Reinke P, Rahmani Mashhour A, Günther S, Niebling S, Schamoni-Kast K, Uetrecht C, Meents A, Chapman HN, Sprenger J, Lane TJ. (2025) Statistical crystallography reveals an allosteric network in SARS-CoV-2 Mpro. bioRxiv https://doi.org/10.1101/2025.01.28.635305
2021
Mischitz V. Editors: Cross A, Wissel S, Uetrecht C, Lorenzen K (2021) Norovirus Superstar (in English or German): Comic book of for the EU project MS SPIDOC. Doi: 10.22003/XFEL.EU-COMIC-2021-001-EN / 10.22003/XFEL.EU-COMIC-2021-001-DE
Kadek A, Lorenzen K, Uetrecht C (2021) In a flash of light: X-ray free electron lasers meet native mass spectrometry. Drug Discov Today Technol. 39:89-99. doi: 10.1016/j.ddtec.2021.07.001.
Konietzny A, Grendel J, Kadek A, Bucher M, Han Y, Hertrich N, Dekkers DHW, Demmers JAA, Grünewald K, Uetrecht C, Mikhaylova M. (2021) Caldendrin and myosin V regulate synaptic spine apparatus localization via ER stabilization in dendritic spines.EMBO J. e106523. doi: 10.15252/embj.2020106523.
Yan H, Lockhauserbäumer J, Szekeres GP, Mallagaray A, Creutznacher R, Taube S, Peters T, Pagel K, Uetrecht C (2021) Protein Secondary Structure Affects Glycan Clustering in Native Mass Spectrometry. Life (Basel). 11(6). doi: 10.3390/life11060554.
Günther S, Reinke PYA, Fernández-García Y, Lieske J, Lane TJ, Ginn HM, Koua FHM, Ehrt C, Ewert W, Oberthuer D, Yefanov O, Meier S, Lorenzen K, Krichel B, Kopicki JD, Gelisio L, Brehm W, Dunkel I, Seychell B, Gieseler H, Norton-Baker B, Escudero-Pérez B, Domaracky M, Saouane S, Tolstikova A, White TA, Hänle A, Groessler M, Fleckenstein H, Trost F, Galchenkova M, Gevorkov Y, Li C, Awel S, Peck A, Barthelmess M, Schlünzen F, Lourdu Xavier P, Werner N, Andaleeb H, Ullah N, Falke S, Srinivasan V, França BA, Schwinzer M, Brognaro H, Rogers C, Melo D, Zaitseva-Doyle JJ, Knoska J, Peña-Murillo GE, Mashhour AR, Hennicke V, Fischer P, Hakanpää J, Meyer J, Gribbon P, Ellinger B, Kuzikov M, Wolf M, Beccari AR, Bourenkov G, von Stetten D, Pompidor G, Bento I, Panneerselvam S, Karpics I, Schneider TR, Garcia-Alai MM, Niebling S, Günther C, Schmidt C, Schubert R, Han H, Boger J, Monteiro DCF, Zhang L, Sun X, Pletzer-Zelgert J, Wollenhaupt J, Feiler CG, Weiss MS, Schulz EC, Mehrabi P, Karničar K, Usenik A, Loboda J, Tidow H, Chari A, Hilgenfeld R, Uetrecht C, Cox R, Zaliani A, Beck T, Rarey M, Günther S, Turk D, Hinrichs W, Chapman HN, Pearson AR, Betzel C, Meents A (2021) X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 372(6542):642-646. doi: 10.1126/science.abf7945.
Simanjuntak Y, Schamoni-Kast K, Grün A, Uetrecht C, Scaturro P (2021) Top-Down and Bottom-Up Proteomics Methods to Study RNA Virus Biology. Viruses.13(4). doi: 10.3390/v13040668.
Dülfer J, Yan H, Brodmerkel MN, Creutznacher R, Mallagaray A, Peters T, Caleman C, Marklund EG, Uetrecht C (2021) Glycan-Induced Protein Dynamics in Human Norovirus P Dimers Depend on Virus Strain and Deamidation Status. Molecules. 26(8). doi: 10.3390/molecules26082125.
Krichel B, Bylapudi G, Schmidt C, Blanchet C, Schubert R, Brings L, Koehler M, Zenobi R, Svergun D, Lorenzen K, Madhugiri R, Ziebuhr J, Uetrecht C (2021) Hallmarks of Alpha- and Betacoronavirus non-structural protein 7+8 complexes. Sci Adv. 7(10). doi: 10.1126/sciadv.abf1004.
Han H, Round E, Schubert R, Gül Y, Makroczyová J, Meza D, Heuser P, Aepfelbacher M, Barák I, Betzel C, Fromme P, Kursula I, Nissen P, Tereschenko E, Schulz J, Uetrecht C, Ulicný J, Wilmanns M, Hajdu J, Lamzin VS, Lorenzen K (2021) The XBI BioLab for life science experiments at the European XFEL. J Appl Crystallogr. 54(Pt 1):7-21. doi: 10.1107/S1600576720013989.
2020
Stanelle-Bertram S, Schaumburg B, Mounogou Kouassi N, Beck S, Zickler M, Beythien G, Becker K, Bai T, Jania H, Müller Z, Pinho dos Reis V, Krump-Buzumkic V, Kadek A, Uetrecht C, Schroeder M, Jarczak D, Nierhaus A, Kluge S, Scaturro P, Baumgärtner W, Klingel K, Gabriel G (2020) SARS-CoV-2 induced CYP19A1 expression in the lung correlates with increased aromatization of testosterone-to-estradiol in male golden hamsters.researchsquare, doi: 10.21203/rs.3.rs-107474/v1
Struwe W, Emmott E, Bailey M, Sharon M, Sinz A, Corrales FJ, Thalassinos K, Braybrook J, Mills C, Barran P (2020) The COVID-19 MS Coalition—accelerating diagnostics, prognostics, and treatment. On behalf of the COVID-19 MS Coalition. Lancet, P1761-1762, https://doi.org/10.1016/S0140-6736(20)31211-3
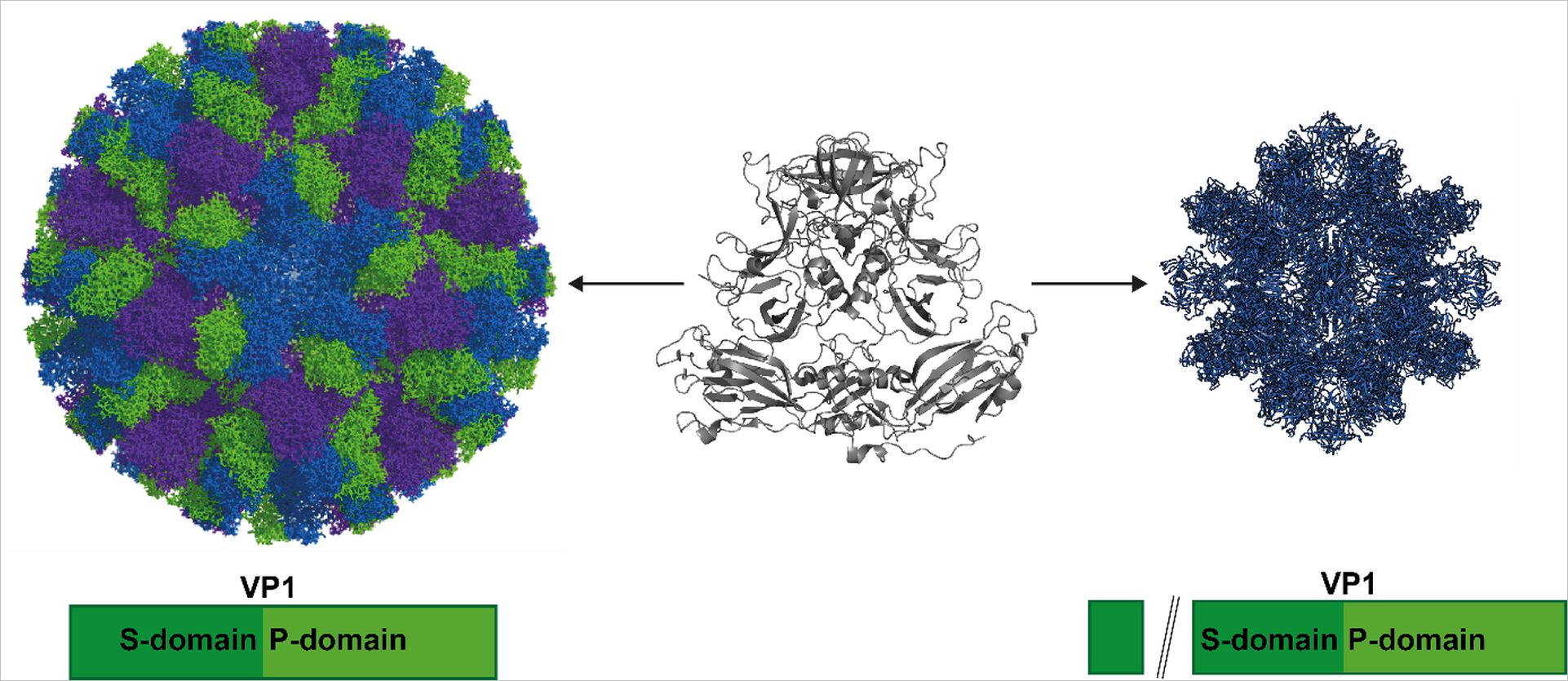
Pogan R, Weiss VU, Bond K, Dülfer J, Krisp C, Lyktey N, Müller-Guhl J, Zoratto S, Allmaier G, Jarrold MF, Muñoz-Fontela C, Schlüter H, Uetrecht C (2020) N-terminal VP1 Truncations Favor T = 1 Norovirus-Like Particles. Vaccines (Basel). 9(1). doi: 10.3390/vaccines9010008.
Schulz EC, Henderson SR, Illarionov B, Crosskey T, Southall SM, Krichel B, Uetrecht C, Fischer M, Wilmanns M (2020) The crystal structure of mycobacterial epoxide hydrolase A. Sci Rep. 2020 Oct 6;10(1):16539. doi: 10.1038/s41598-020-73452-y.
Sobolev E, Zolotarev S, Giewekemeyer K, Bielecki J, Okamoto K, Reddy H, Andreasson J, Ayyer K, Barak I, Bari S, Barty A, Bean R, Bobkov S, Chapman H, Chojnowski G, Daurer B, Dörner K, Ekeberg T, Flückiger L, Galzitskaya O, Gelisio L, Hauf S, Hogue B, Horke D, Hosseinizadeh A, Ilyin V, Jung C, Kim C, Kim Y, Kirian R, Kirkwood H, Kulyk O, Küpper J, Letrun R, Loh N, Lorenzen K, Messerschmidt M, Mühlig K, Ourmazd A, Raab N, Rode A, Rose M, Round A, Sato T, Schubert R, Schwander P, Sellberg J, Sikorski M, Silenzi A, Song C, Spence J, Stern S, Sztuk-Dambietz J, Teslyuk A, Timneanu N, Trebbin M, Uetrecht C, Weinhausen B, Williams G, Xavier P, Xu C, Vartanyants I, Lamzin V, Mancuso A, Maia F (2020) Megahertz single-particle imaging at the European XFEL. Communications Physics. 3(1): doi: 10.1038/s42005-020-0362-y.
Günther S, Reinke P, Oberthuer D, Yefanov O, Ginn H, Meier S, Lane T, Lorenzen K, Gelisio L, Brehm W, Dunkel I, Domaracky M, Saouane S, Lieske J, Ehrt C, Koua F, Tolstikova A, White T, Groessler M, Fleckenstein H, Trost F, Galchenkova M, Gevorkov Y, Li C, Awel S, Peck A, Xavier P, Barthelmess M, Schlünzen F, Werner N, Andaleeb H, Ullah N, Falke S, Franca B, Schwinzer M, Brognaro H, Seychell B, Gieseler H, Melo D, Zaitsev-Doyle J, Norton-Baker B, Knoska J, Esperanza G, Mashhour A, Guicking F, Hennicke V, Fischer P, Rogers C, Monteiro D, Hakanpää J, Meyer J, Noei H, Gribbon P, Ellinger B, Kuzikov M, Wolf M, Zhang L, Sun X, Pletzer-Zelgert J, Wollenhaupt J, Feiler C, Weiss M, Schulz E, Mehrabi P, Schmidt C, Schubert R, Han H, Krichel B, Fernández-García Y, Escudero-Pérez B, Günther S, Turk D, Uetrecht C, Beck T, Tidow H, Chari A, Zaliani A, Rarey M, Cox R, Hilgenfeld R, Chapman H, Pearson A, Betzel C, Meents A (2020) Catalytic cleavage of HEAT and subsequent covalent binding of the tetralone moiety by the SARS-CoV-2 main protease. bioRxiv. doi: 10.1101/2020.05.02.043554.
Horstmann JA, Lunelli M, Cazzola H, Heidemann J, Kühne C, Steffen P, Szefs S, Rossi C, Lokareddy RK, Wang C, Lemaire L, Hughes KT, Uetrecht C, Schlüter H, Grassl GA, Stradal TEB, Rossez Y, Kolbe M, Erhardt M (2020) Methylation of Salmonella Typhimurium flagella promotes bacterial adhesion and host cell invasion. Nat Commun. 11(1):2013. doi: 10.1038/s41467-020-15738-3.
Krichel B, Falke S, Hilgenfeld R, Redecke L, Uetrecht C (2020) Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem J. 477(5):1009-1019. doi: 10.1042/BCJ20200029.
Anjanappa R, Garcia-Alai M, Kopicki JD, Lockhauserbäumer J, Aboelmagd M, Hinrichs J, Nemtanu IM, Uetrecht C, Zacharias M, Springer S, Meijers R (2020) Structures of peptide-free and partially loaded MHC class I molecules reveal mechanisms of peptide selection. Nat Commun. 11(1):1314. doi: 10.1038/s41467-020-14862-4.
Heidemann J, Kölbel K, Konijnenberg A, Van Dyck J, Garcia-Alai M, Meijers R, Sobott F, Uetrecht C (2020) Further insights from structural mass spectrometry into endocytosis adaptor protein assemblies. International Journal of Mass Spectrometry. 447:116240-. doi: 10.1016/j.ijms.2019.116240.
2019
Bernal I, Römermann J, Flacht L, Lunelli M, Uetrecht C, Kolbe M (2019) Structural analysis of ligand-bound states of the Salmonella type III secretion system ATPase InvC. Protein Sci. 28(10):1888-1901. doi: 10.1002/pro.3704.
Bernal I, Börnicke J, Heidemann J, Svergun D, Horstmann JA, Erhardt M, Tuukkanen A, Uetrecht C, Kolbe M (2019) Molecular Organization of Soluble Type III Secretion System Sorting Platform Complexes. J Mol Biol. 431(19):3787-3803. doi: 10.1016/j.jmb.2019.07.004.
Weiss VU, Pogan R, Zoratto S, Bond KM, Boulanger P, Jarrold MF, Lyktey N, Pahl D, Puffler N, Schelhaas M, Selivanovitch E, Uetrecht C, Allmaier G (2019) Virus-like particle size and molecular weight/mass determination applying gas-phase electrophoresis (native nES GEMMA). Anal Bioanal Chem. 411(23):5951-5962. doi: 10.1007/s00216-019-01998-6.
Dülfer J, Kadek A, Kopicki JD, Krichel B, Uetrecht C (2019) Structural mass spectrometry goes viral. Adv Virus Res. 105:189-238. doi: 10.1016/bs.aivir.2019.07.003.
Kesgin-Schaefer S, Heidemann J, Puchert A, Koelbel K, Yorke BA, Huse N, Pearson AR, Uetrecht C, Tidow H (2019) Crystal structure of a domain-swapped photoactivatable sfGFP variant provides evidence for GFP folding pathway. FEBS J. 286(12):2329-2340. doi: 10.1111/febs.14797
Uetrecht C, Lorenzen K, Kitel M, Heidemann J, Robinson Spencer JH, Schlüter H, Schulz J (2019) Native mass spectrometry provides sufficient ion flux for XFEL single-particle imaging. J Synchrotron Radiat. 26(Pt 3):653-659. doi: 10.1107/S1600577519002686.
Mallagaray A, Creutznacher R, Dülfer J, Mayer PHO, Grimm LL, Orduña JM, Trabjerg E, Stehle T, Rand KD, Blaum BS, Uetrecht C, Peters T (2019) A post-translational modification of human Norovirus capsid protein attenuates glycan binding. Nat Commun. 10(1):1320. doi: 10.1038/s41467-019-09251-5.
Zaitsev-Doyle J, Puchert A, Pfeifer Y, Yan H, Yorke B, Müller-Werkmeister H, Uetrecht C, Rehbein J, Huse N, Pearson A, Sans M (2019) Synthesis and characterisation of α-carboxynitrobenzyl photocaged l -aspartates for applications in time-resolved structural biology. RSC Advances. 9(15):8695-8699. doi: 10.1039/C9RA00968J.
2018
Heidemann J, Krichel B, Uetrecht C. (2018) Native Massenspektrometrie für die Proteinstrukturanalytik. BIOspektrum, 24:164-167.
Holm T, Kopicki JD, Busch C, Olschewski S, Rosenthal M, Uetrecht C, Günther S, Reindl S (2018) Biochemical and structural studies reveal differences and commonalities among cap-snatching endonucleases from segmented negative-strand RNA viruses. J Biol Chem. 293(51):19686-19698. doi: 10.1074/jbc.RA118.004373.
Nitsche J, Josts I, Heidemann J, Mertens HD, Maric S, Moulin M, Haertlein M, Busch S, Forsyth VT, Svergun DI, Uetrecht C, Tidow H (2018) Structural basis for activation of plasma-membrane Ca2+-ATPase by calmodulin. Commun Biol. 1:206. doi: 10.1038/s42003-018-0203-7.
Bücher KS, Yan H, Creutznacher R, Ruoff K, Mallagaray A, Grafmüller A, Dirks JS, Kilic T, Weickert S, Rubailo A, Drescher M, Schmidt S, Hansman G, Peters T, Uetrecht C, Hartmann L (2018) Fucose-Functionalized Precision Glycomacromolecules Targeting Human Norovirus Capsid Protein. Biomacromolecules. 19(9):3714-3724. doi: 10.1021/acs.biomac.8b00829.
Pogan R, Dülfer J, Uetrecht C (2018) Norovirus assembly and stability. Curr Opin Virol. 31:59-65. doi: 10.1016/j.coviro.2018.05.003.
Shin HC, Deterra D, Park J, Kim H, Nishikiori M, Uetrecht C, Ahlquist PG, Arbulu M, Blick RH (2018) Ultra-high mass multimer analysis of protein-1a capping domains by a silicon nanomembrane detector. J Proteomics. 175:5-11. doi: 10.1016/j.jprot.2017.11.024.
Pogan R, Schneider C, Reimer R, Hansman G, Uetrecht C (2018) Norovirus-like VP1 particles exhibit isolate dependent stability profiles. J Phys Condens Matter. 30(6):064006. doi: 10.1088/1361-648X/aaa43b.
Garcia-Alai MM, Heidemann J, Skruzny M, Gieras A, Mertens HDT, Svergun DI, Kaksonen M, Uetrecht C, Meijers R (2018) Epsin and Sla2 form assemblies through phospholipid interfaces. Nat Commun. 9(1):328. doi: 10.1038/s41467-017-02443-x
2017
Wegener H, Mallagaray Á, Schöne T, Peters T, Lockhauserbäumer J, Yan H, Uetrecht C, Hansman GS, Taube S (2017) Human norovirus GII.4(MI001) P dimer binds fucosylated and sialylated carbohydrates. Glycobiology. 27(11):1027-1037. doi: 10.1093/glycob/cwx078
2016
Szameit K, Berg K, Kruspe S, Valentini E, Magbanua E, Kwiatkowski M, Chauvot de Beauchêne I, Krichel B, Schamoni K, Uetrecht C, Svergun DI, Schlüter H, Zacharias M, Hahn U (2016) Structure and target interaction of a G-quadruplex RNA-aptamer. RNA Biol. 13(10):973-987. doi: 10.1080/15476286.2016.1212151.
Snijder J, Kononova O, Barbu IM, Uetrecht C, Rurup WF, Burnley RJ, Koay MS, Cornelissen JJ, Roos WH, Barsegov V, Wuite GJ, Heck AJ (2016) Assembly and Mechanical Properties of the Cargo-Free and Cargo-Loaded Bacterial Nanocompartment Encapsulin. Biomacromolecules. 17(8):2522-9. doi: 10.1021/acs.biomac.6b00469.
Hantke MF, Hasse D, Ekeberg T, John K, Svenda M, Loh D, Martin AV, Timneanu N, Larsson DS, van der Schot G, Carlsson GH, Ingelman M, Andreasson J, Westphal D, Iwan B, Uetrecht C, Bielecki J, Liang M, Stellato F, DePonte DP, Bari S, Hartmann R, Kimmel N, Kirian RA, Seibert MM, Mühlig K, Schorb S, Ferguson K, Bostedt C, Carron S, Bozek JD, Rolles D, Rudenko A, Foucar L, Epp SW, Chapman HN, Barty A, Andersson I, Hajdu J, Maia FR (2016) A data set from flash X-ray imaging of carboxysomes. Sci Data. 3:160061. doi: 10.1038/sdata.2016.61.
Dunne M, Leicht S, Krichel B, Mertens HD, Thompson A, Krijgsveld J, Svergun DI, Gómez-Torres N, Garde S, Uetrecht C, Narbad A, Mayer MJ, Meijers R (2016) Crystal Structure of the CTP1L Endolysin Reveals How Its Activity Is Regulated by a Secondary Translation Product. J Biol Chem. 291(10):4882-93. doi: 10.1074/jbc.M115.671172.
2015
Mallagaray A, Lockhauserbäumer J, Hansman G, Uetrecht C, Peters T (2015) Attachment of norovirus to histo blood group antigens: a cooperative multistep process. Angew Chem Int Ed Engl. 54(41):12014-9. doi: 10.1002/anie.201505672.
von der Heyde A, Lockhauserbäumer J, Uetrecht C, Elleuche S (2015) A hydrolase-based reporter system to uncover the protein splicing performance of an archaeal intein. Appl Microbiol Biotechnol. 99(18):7613-24. doi: 10.1007/s00253-015-6689-8.
Skruzny M, Desfosses A, Prinz S, Dodonova SO, Gieras A, Uetrecht C, Jakobi AJ, Abella M, Hagen WJ, Schulz J, Meijers R, Rybin V, Briggs JA, Sachse C, Kaksonen M (2015) An organized co-assembly of clathrin adaptors is essential for endocytosis. Dev Cell. 33(2):150-62. doi: 10.1016/j.devcel.2015.02.023.
2013
Tseng YH, Uetrecht C, Yang SC, Barendregt A, Heck AJ, Peng WP (2013) Game-theory-based search engine to automate the mass assignment in complex native electrospray mass spectra. Anal Chem. 85(23):11275-83. doi: 10.1021/ac401940e.
Snijder J, Uetrecht C, Rose RJ, Sanchez-Eugenia R, Marti GA, Agirre J, Guérin DM, Wuite GJ, Heck AJ, Roos WH (2013) Probing the biophysical interplay between a viral genome and its capsid. Nat Chem. 5(6):502-9. doi: 10.1038/nchem.1627.
Broeker NK, Gohlke U, Müller JJ, Uetrecht C, Heinemann U, Seckler R, Barbirz S (2013) Single amino acid exchange in bacteriophage HK620 tailspike protein results in thousand-fold increase of its oligosaccharide affinity. Glycobiology. 23(1):59-68. doi: 10.1093/glycob/cws126.
Schulz J, Bari S, Buck J, Uetrecht C (2013) Sample refreshment schemes for high repetition rate FEL experiments. In: Tschentscher T, Tiedtke K, editors. SPIE Optics + Optoelectronics
2012
Zaccheus MV, Broeker NK, Lundborg M, Uetrecht C, Barbirz S, Widmalm G (2012) Structural studies of the O-antigen polysaccharide from Escherichia coli TD2158 having O18 serogroup specificity and aspects of its interaction with the tailspike endoglycosidase of the infecting bacteriophage HK620. Carbohydr Res. 357:118-25. doi: 10.1016/j.carres.2012.05.022.
2011
Baclayon M, Shoemaker GK, Uetrecht C, Crawford SE, Estes MK, Prasad BV, Heck AJ, Wuite GJ, Roos WH (2011) Prestress strengthens the shell of Norwalk virus nanoparticles. Nano Lett. 11(11):4865-9. doi: 10.1021/nl202699r.
Uetrecht C, Heck AJ (2011) Modern biomolecular mass spectrometry and its role in studying virus structure, dynamics, and assembly. Angew Chem Int Ed Engl. 50(36):8248-62. doi: 10.1002/anie.201008120.
Brasch M, de la Escosura A, Ma Y, Uetrecht C, Heck AJ, Torres T, Cornelissen JJ (2011) Encapsulation of phthalocyanine supramolecular stacks into virus-like particles. J Am Chem Soc. 133(18):6878-81. doi: 10.1021/ja110752u.
Tseng YH, Uetrecht C, Heck AJ, Peng WP (2011) Interpreting the charge state assignment in electrospray mass spectra of bioparticles. Anal Chem. 83(6):1960-8. doi: 10.1021/ac102676z.
Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, Heck AJ (2011) Interrogating viral capsid assembly with ion mobility-mass spectrometry. Nat Chem. 3(2):126-32. doi: 10.1038/nchem.947.
Karagöz GE, Duarte AM, Ippel H, Uetrecht C, Sinnige T, van Rosmalen M, Hausmann J, Heck AJ, Boelens R, Rüdiger SG (2011) N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc Natl Acad Sci U S A. 108(2):580-5. doi: 10.1073/pnas.1011867108.
2010
Uetrecht C, Watts NR, Stahl SJ, Wingfield PT, Steven AC, Heck AJ (2010) Subunit exchange rates in Hepatitis B virus capsids are geometry- and temperature-dependent. Phys Chem Chem Phys12(41):13368-71. doi: 10.1039/c0cp00692k.
Roos WH, Gibbons MM, Arkhipov A, Uetrecht C, Watts NR, Wingfield PT, Steven AC, Heck AJ, Schulten K, Klug WS, Wuite GJ (2010) Squeezing protein shells: how continuum elastic models, molecular dynamics simulations, and experiments coalesce at the nanoscale. Biophys J. 99(4):1175-81. doi: 10.1016/j.bpj.2010.05.033.
Fu CY, Uetrecht C, Kang S, Morais MC, Heck AJ, Walter MR, Prevelige PE Jr (2010) A docking model based on mass spectrometric and biochemical data describes phage packaging motor incorporation. Mol Cell Proteomics. 1764-73. doi: 10.1074/mcp.M900625-MCP200.
Shoemaker GK, van Duijn E, Crawford SE, Uetrecht C, Baclayon M, Roos WH, Wuite GJ, Estes MK, Prasad BV, Heck AJ (2010) Norwalk virus assembly and stability monitored by mass spectrometry. Mol Cell Proteomics. 1742-51. doi: 10.1074/mcp.M900620-MCP200.
Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJ (2010) Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 39(5):1633-55. doi: 10.1039/b914002f.
2008
Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJ, Wingfield PT, Steven AC, Heck AJ (2008) High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc Natl Acad Sci U S A. 105(27):9216-20. doi: 10.1073/pnas.0800406105.
Barbirz S, Müller JJ, Uetrecht C, Clark AJ, Heinemann U, Seckler R (2008) Crystal structure of Escherichia coli phage HK620 tailspike: podoviral tailspike endoglycosidase modules are evolutionarily related. Mol Microbiol. 69(2):303-16. doi: 10.1111/j.1365-2958.2008.06311.x.
Lorenzen K, Olia AS, Uetrecht C, Cingolani G, Heck AJ (2008) Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J Mol Biol. 379(2):385-96. doi: 10.1016/j.jmb.2008.02.017.
Uetrecht C, Versluis C, Watts NR, Wingfield PT, Steven AC, Heck AJ (2008) Stability and shape of hepatitis B virus capsids in vacuo. Angew Chem Int Ed Engl. 47(33):6247-51. doi: 10.1002/anie.200802410.