Key Virulence Factor of Salmonella Characterized
CSSB's Kolbe group and collaborators reveal that the flagellin protein, FliB, initiates an important reaction in Salmonella enterica which is mediated by an iron-sulfur cluster.
Salmonella enterica, a foodborne pathogen, is 1 of 4 key global causes of diarrheal diseases which affect over 550 million people each year. This rod-shaped, Gram-negative bacterium has tubular, whip-like appendages known as flagella that protrude from its surface. The flagella, which is comprised of tens of thousands of flagellin proteins, is not only important for the movement of the bacteria but also plays a key role in its ability to invade the human host cell.
The parts of the flagellin proteins that are exposed are highly versatile and subject to modification by enzymes. One such modification, known as methylation, occurs when a methyl group is added to a protein thus giving the protein the ability to function. Protein methylation plays an integral role in physiological function and signaling pathway modulation in Salmonella enterica.
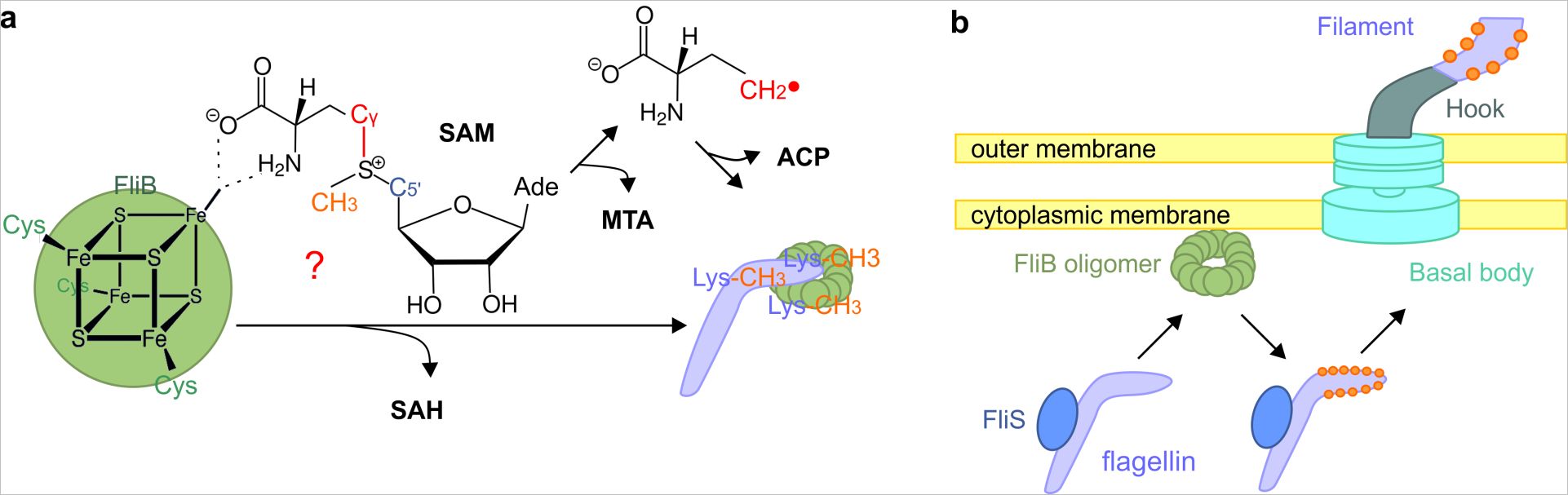
In the flagella, residues of the amino acid lysine on the flagellin outer surface are methylated to ɛ-N-methyl-lysine. In a previous paper, the researchers showed the flagellin’s importance in host cell innovation by demonstrating that flagellin methylation facilitates the adhesion of Salmonella to host cell surfaces. Although it was the first methylase ever discovered more than fifty years ago, little is known about FliB the methylase responsible for catalyzing this process
Researchers from the Kolbe group in collaboration with researchers at CSSB’s newest partner institution, Research Center Borstel FZB, as well as Max Plank Institute for Infection Biology, the University of Marburg and the Technical University of Braunschweig have revealed that FliB catalyzes a methyl transfer reaction which is mediated by an iron-sulfur cluster, [4Fe-4S]. These insights into FliB’s biochemical and biophysical properties were recently published in PLOS Pathogens. According to the researchers, their study sets the stage for understanding the roles of iron-sulfer clusters and radical chemistry in flagellar modification.
The researchers were able demonstrate that FliB is a S-adenosylmethionine dependent methyltransferase. ‘We also observed that FliB is located in the bacterial cytoplasm,” explains the paper’s first author Chu Wang “this indicates methylation happens prior to flagellin secretion and flagellar assembly.”
To examine FliB in more detail the researchers established a new procedure to purify FliB by creating a genetic construct which fuses FliB it to the end of a flagellin protein from a different Gram-negative bacterium. This genetic construct, known as ftFilB, could then be examined using a several different methods including single particle EM, X-ray fluorescence, Mößbauer spectroscopy and atomic and atomic force microscopy.
The researchers used the new Atomic Force microscope, recently installed at CSSB by the FZB group of Thomas Gutsmann, to measure the diameter and height of the tFilB complex. “We were able to determine that ftFliB forms a spherical complex with a molecular mass in the Megalton range,” explains Gutsmann.
“The new insights generated into FliB have helped us characterize a key virulence factor of Salmonella enterica,” explains Michael Kolbe the paper’s corresponding author “Although, FliB has a low sequence conservation, it is present in many, many different bacterial pathogens and could therefore be an interesting target for future studies.”
Source:
Wang C, Nehls C, Baabe D, Burghaus O, Hurwitz R, Gutsmann T, Bröring M, Kolbe M. (2021) Flagellin lysine methyltransferase FliB catalyzes a [4Fe-4S] mediated methyl transfer reaction. PLoS Pathog.17(11):e1010052. doi: 10.1371/journal.ppat.1010052
*Teaser Image: CDC/ Antibiotic Resistance Coordination and Strategy Unit, Medical Illustrator: James Archer



