The final step in RNA degradation: New insights into bacterial dinucleotidases
Based on a new structure-function study the Sondermann group (DESY) in collaboration with groups at the University of Maryland and Cornell University (USA) defined the function of NrnC-type RNases, essential enzymes that catalyze the last step of RNA degradation in many bacteria.
RNA is a versatile biopolymer, which is responsible for the coding, regulation, and expression of genes within a cell. Many RNAs have relatively short lifespans, enabling for example the regulation of gene expression via RNA degradation. During RNA degradation various classes of enzymes work together to break down an RNA polymer into smaller molecules. In the final steps, small, 2-5 nucleotide-long fragments are degraded by specialized enzymes, so-called oligoribonucleases or nano-RNases. Sondermann revealed that oligoribonuclease (‘Orn’ in short), unlike its name would suggest and previous descriptions have shown, likely acts as a dedicated dinucleotidase with a very strong preference for RNAs with only two nucleotides, thus catalyzing the final step in RNA degradation (Kim et al., 2019). The current study builds on this discovery, demonstrating that another nano-RNase, NrnC, shares this feature with Orn – despite their independent evolution.
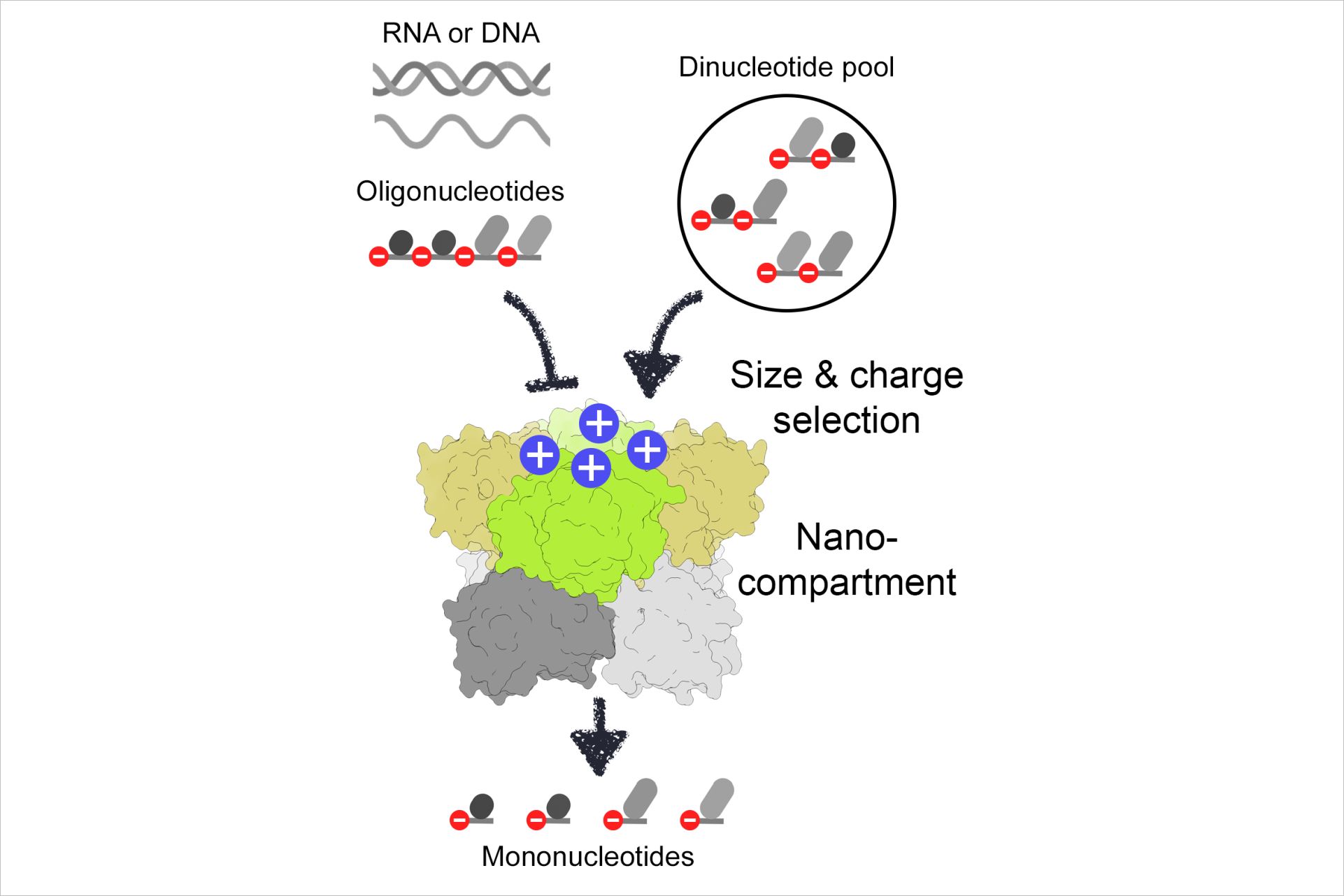
Nano-RNase C (NrnC) is present in many organisms that lack Orn. NrnC can also stand in for Orn in functional assays, suggesting overlapping functions. How NrnC would selectively degrade only RNAs 2-5 nucleotides in length, but less so longer substrates had remained enigmatic. In a recent publication in eLife, the researchers provide strong evidence that NrnC, just like Orn, has a much higher preference for dinucleotides than assumed previously. Structural studies using X-ray crystallography and cryo-electron microscopy revealed a narrow active site of NrnC with a catalytically competent state only observed with dinucleotides. Quantitative binding and activity assays support this notion regarding NrnC’s substrate preference. “We discovered that at least two enzymes, NrnC and Orn, degrade specifically diribonucleotides into mono-ribonucleotides in the final step of RNA decay,” explains Holger Sondermann. “Interestingly, the two enzyme families appear to have evolved independently arriving at two solutions to resolve the cellular dinucleotide pool.”
Of note, the two enzymes, NrnC and Orn, are essential in many organisms, suggesting that the buildup of dinucleotides is somehow toxic for cells. The similar function yet evolutionary distinct histories of NrnC and Orn indicate that dinucleotidase activity is necessary for proper cellular growth. “Fully understanding how NrnC functions could offer novel strategies to combat gram-negative pathogens in infectious disease,” notes Sondermann.
The work was supported by the National Institutes of Health (USA). It is based on a close collaboration between the groups of Drs. Vincent T. Lee and Wade C. Winkler at the University of Maryland, USA. Other authors include Soo-Kyoung Kim, George A. Walters-Marrah, Bryce A. Brownfield, J. Christopher Fromme, and Jonathan R. Goodson.
Sources:
Lormand JD, Kim SK, Walters-Marrah GA, Brownfield BA, Fromme JC, Winkler WC, Goodson JR, Lee VT, Sondermann H. (2021) Structural characterization of NrnC identifies unifying features of dinucleotidases. Elife. 10:e70146. doi: 10.7554/eLife.70146.
Kim SK, Lormand JD, Weiss CA, Eger KA, Turdiev H, Turdiev A, Winkler WC, Sondermann H, Lee VT (2019) A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation. Elife 810.7554/eLife.46313 doi: 10.7554/eLife.46313



