Mapping Atoms
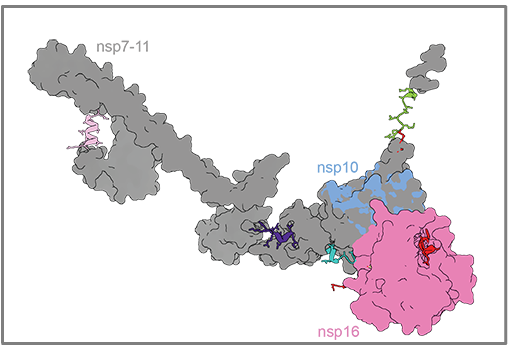
The Marlovits group specializes in building detailed, high-resolution models of molecular machines such as the Type VII secretion system and the RuvAB branch migration complex. To facilitate the building of these models, the group developed StarMap, an easy-to-use software program for molecular refinements using Rosetta. A description of the StarMap software package was recently published in Nature Protocols.
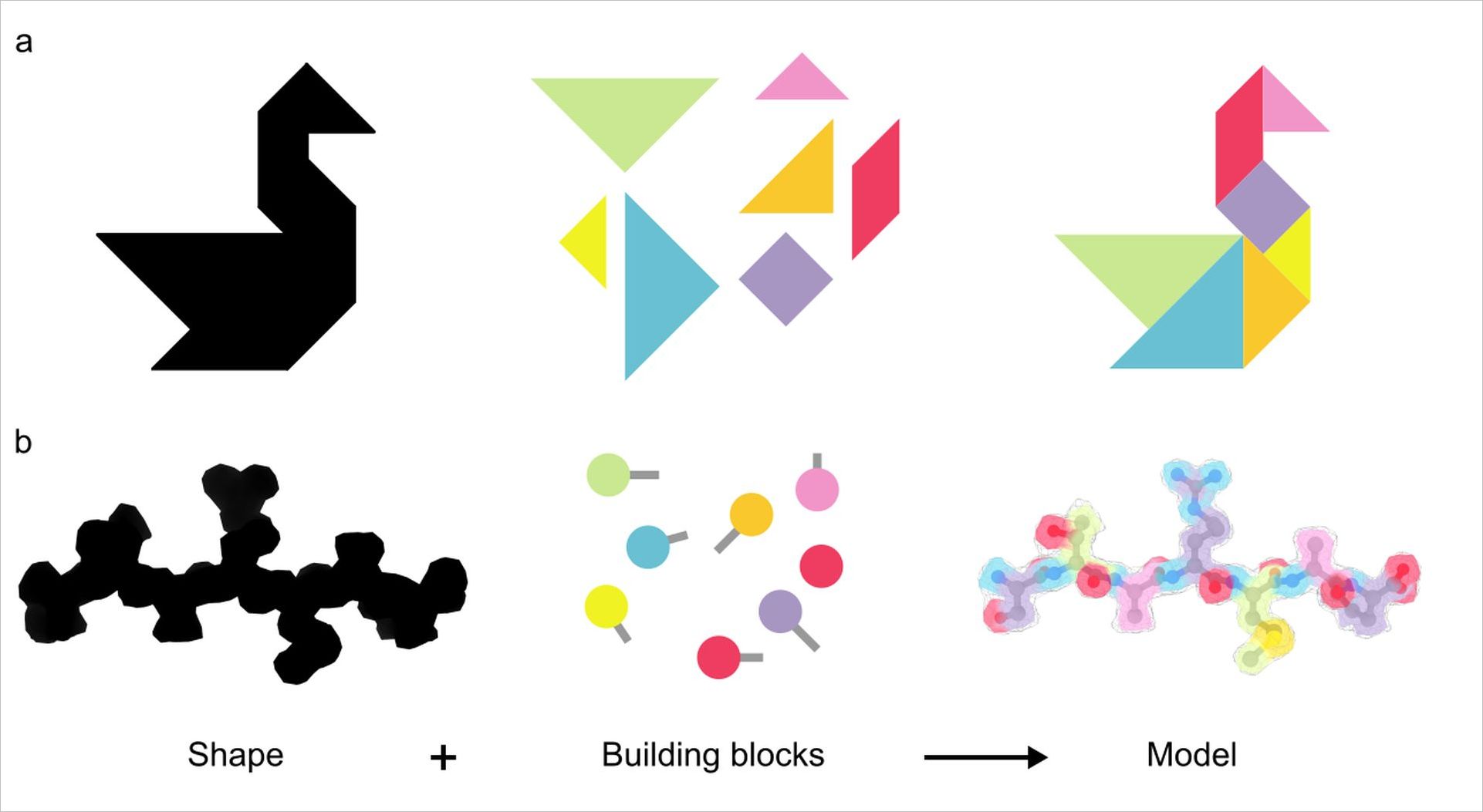
Composed of thousands of atoms that interact with one another in a precise manner molecular machines are unbelievably intricate and complex. The paper’s first authors Wolfgang Lugmayr and Vadim Kotov compared the task of building a model of these molecular machines to solving a tangram puzzle. In their “behind the paper” blog post they note “you build a (tangram) object by combining simple shapes like squares and triangles (Figure 1a). In structural biology the goal is to represent the 3-dimensional shape of a molecule with spheres (atoms) and sticks (bonds) as depicted in Figure 1b.”
Unlike a tangram, however, there is more than one way to “solve” a molecular model and this is where StarMap is able to help. StarMap is an easy-to-use interface, between the popular structural display program ChimeraX and the powerful molecular modeling engine Rosetta, that provides users with a general approach for refining structural models. “Just as astronomers use star maps to pinpoint the exact location of a star in the night sky, our StarMap software helps to reveal the precise location of atoms in a molecular model,” notes Marlovits.
Unlike some other refinement programs, StarMap users do not need to have coding expertise to build models with atomic resolution. Given its ease-of-use and accuracy, the Marlovits group believes that StarMap will soon become an essential tool for scientists using cryo-EM to investigate complex molecular machines.
Behind the Paper:
https://protocolsmethods.springernature.com/posts/how-to-solve-a-molecular-tangram
Source:
Lugmayr W, Kotov V, Goessweiner-Mohr N, Wald J, DiMaio F, Marlovits TC (2022) StarMap: a user-friendly workflow for Rosetta-driven molecular structure refinement. Nat Protoc. doi: 10.1038/s41596-022-00757-9.